













实验动物与比较医学 ›› 2023, Vol. 43 ›› Issue (5): 548-558.DOI: 10.12300/j.issn.1674-5817.2023.073
赵丽亚1,2( ), 倪丽菊1(
), 倪丽菊1( ), 张彩勤3, 汤建平1,2, 姚养正4, 聂艳艳1, 顾晓雪2, 赵莹1(
), 张彩勤3, 汤建平1,2, 姚养正4, 聂艳艳1, 顾晓雪2, 赵莹1( )(
)( )
)
收稿日期:2023-06-09
修回日期:2023-08-08
出版日期:2023-10-25
发布日期:2023-10-25
通讯作者:
赵 莹(1982—),女,硕士,副研究员,研究方向:实验动物遗传。E-mail: zhaoying@slarc.org.cn。ORCID: 0000-0002-0551-0566作者简介:赵丽亚(1981—),女,硕士,工程师,研究方向:分子遗传学。E-mail: Zhaoliya3002@163.com;
倪丽菊(1979—),女,硕士,副研究员,研究方向:实验动物遗传。E-mail: niliju@163.com
Liya ZHAO1,2( ), Liju NI1(
), Liju NI1( ), Caiqin ZHANG3, Jianping TANG1,2, Yangzheng YAO4, Yanyan NIE1, Xiaoxue GU2, Ying ZHAO1(
), Caiqin ZHANG3, Jianping TANG1,2, Yangzheng YAO4, Yanyan NIE1, Xiaoxue GU2, Ying ZHAO1( )(
)( )
)
Received:2023-06-09
Revised:2023-08-08
Published:2023-10-25
Online:2023-10-25
Contact:
ZHAO Ying (ORCID: 0000-0002-0551-0566), E-mail: zhaoying@slarc.org.cn摘要:
目的 建立一套基于多重PCR-连接酶检测反应(ligase detection reaction,LDR)技术的近交系大鼠单核苷酸多态性(single nucleotide polymorphisms,SNP)检测方案。 方法 在5个品系的SPF级近交系大鼠1~20号常染色体和X染色体上共选取40个大鼠SNP位点,将SNP位点随机分为4组,构建基于多重PCR-LDR技术的近交系大鼠4组SNP位点基因检测方案。采用本方案检测国内另两家大鼠供应商的9个常用大鼠品系。最后,通过第三方实验室对不同DNA聚合酶的扩增效果进行比对,验证本方案的可行性。 结果 用所构建的近交系大鼠SNP遗传检测方案测试5个大鼠品系时,各样本的所有位点均得到了良好的扩增结果。采用本方案检测国内另两家大鼠供应商的9个常用大鼠品系时也得到了良好的扩增结果,40个SNP位点在每个近交系大鼠中均为纯合。用3种来源不同的DNA聚合酶同时检测相同大鼠DNA样本的结果显示,Multiplex PCR Kit、AmpliTaq Gold? 360 DNA聚合酶、PlatinumⅡ Taq热启动DNA聚合酶在第1~3组SNP位点均有扩增产物的电泳峰,其中PlatinumⅡ Taq热启动DNA聚合酶在第4组SNP位点中少了一个扩增产物的电泳峰。另外,不同实验室间的比对结果显示,相同扩增体系的检测结果一致。 结论 基于多重PCR-LDR技术成功建立了一套覆盖所有常染色体与X染色体的大鼠SNP检测方案,该方法的稳定性和重复性俱佳。
中图分类号:
赵丽亚,倪丽菊,张彩勤,等. 基于多重PCR-LDR技术建立近交系大鼠单核苷酸多态性遗传检测方案[J]. 实验动物与比较医学, 2023, 43(5): 548-558. DOI: 10.12300/j.issn.1674-5817.2023.073.
Liya ZHAO,Liju NI,Caiqin ZHANG,et al. Establishing a Genetic Detection Protocol of Single Nucleotide Polymorphisms Panels in Inbred Rats Based on Multiplex PCR-LDR[J]. Laboratory Animal and Comparative Medicine, 2023, 43(5): 548-558. DOI: 10.12300/j.issn.1674-5817.2023.073.
序号 No. | SNP位点 Reference SNP ID | 染色体 Chromosome | 引物名称 Primer name | 引物序列 Primer sequences |
|---|---|---|---|---|
| a1 | rs65970293 | chr1 | rs65970293-up | TGCACATTGGGACTGCTTTA |
| rs65970293-low | ATCTGGCATGGACAGAGCTT | |||
| a2 | rs106996961 | chr2 | rs106996961-up | ATAGGGCCAAGGTCCCATTC |
| rs106996961-low | GAAAAGAACGCTAATCACAGGACA | |||
| a3 | rs63860341 | chr3 | rs63860341-up | CAATGAATCAAAGGCAACCA |
| rs63860341-low | CCACGGTCCCTATGTGAACT | |||
| a4 | rs64750237 | chr4 | rs64750237-up | CAGGTATGGCTTTGGAGGAA |
| rs64750237-low | GTTTTCAGCTGGAGCTGTCC | |||
| a5 | rs107261116 | chr5 | rs107261116-up | CAGCATTGGCAAGTCTTCCT |
| rs107261116-low | TGATAAAAGGAGCAGCAGCA | |||
| a6 | rs64102787 | chr6 | rs64102787-up | CGGGGATCAAAGAGACAAAA |
| rs64102787-low | ATAGTGCCAAAGCCCAACAC | |||
| a7 | rs105187832 | chr7 | rs105187832-up | AGACTACTGGGCTGCTCACC |
| rs105187832-low | CATGGCACACCTTTCTTTCA | |||
| a8 | rs64682441 | chr9 | rs64682441-up | GCTGGAAGTCTCTGGAGTGG |
| rs64682441-low | ACAAGTGGACTGGCTCAGGT | |||
| a9 | rs65067166 | chr10 | rs65067166-up | ATCCCTCTCTTTGCCAGGAT |
| rs65067166-low | GTAGGGTGAGGCTGCTTCTG | |||
| a10 | rs8167985 | chr11 | rs8167985-up | ACCCCCTCTTGCTTTCTGTT |
| rs8167985-low | CTGCTCTTGGATGAAAACCA | |||
| b1 | rs106672258 | chr12 | rs106672258-up | GCCCTTCCCAAGGTCATACT |
| rs106672258-low | AAAGCACAAAAATGCCATCC | |||
| b2 | rs65921305 | chr13 | rs65921305-up | AACCTGGTGATTTGTCAGAGC |
| rs65921305-low | GGCAGACTCCAGGAAGATTG | |||
| b3 | rs106458396 | chr15 | rs106458396-up | AGAGCAGCTGCCAAAATAGC |
| rs106458396-low | AAACCTGGCTTTAGCAAGCA | |||
| b4 | rs106888236 | chr16 | rs106888236-up | TGTGAAGAACACAGGCTCTCC |
| rs106888236-low | TGCCAAACACCATGTAATGAA | |||
| b5 | rs63814610 | chr17 | rs63814610-up | TAGGACAGGGGGAACAACTG |
| rs63814610-low | CCCAAGCATCCTTGACTGTT | |||
| b6 | rs8152541 | chr18 | rs8152541-up | CGTGTCATTCCTGACTGTGG |
| rs8152541-low | ACAGGAGCTGGCACTGAATC | |||
| b7 | rs105824121 | chr19 | rs105824121-up | TTCTGCAATCAAGGGTGGAT |
| rs105824121-low | GGCCACCAAAGATACACAGG | |||
| b8 | rs13453252 | chr20 | rs13453252-up | TACGCCATTTTGCGACATTA |
| rs13453252-low | GGTTCCCTACGAGGAACGAC | |||
| b9 | rs65108991 | chrX | rs65108991-up | CATCCCAGAAGCCCTGTTTA |
| rs65108991-low | GAAGTCAGTGCTGTGCCTCA | |||
| c1 | rs106891788 | chr1 | rs106891788-up | TCCATCTGCTTGGCCCTAAT |
| rs106891788-low | GTTGCAACCCACAGGTTGAG | |||
| c2 | rs107493604 | chr2 | rs107493604-up | TCCTCCTGGAAGACAGCAAC |
| rs107493604-low | GTTCCTGAGAAGGGGCTCTC | |||
| c3 | rs65632519 | chr3 | rs65632519-up | CCCTGTTTCAGCCTTTTGTG |
| rs65632519-low | GAGGAAGAGGGAAGGGAGAA | |||
| c4 | rs107163178 | chr4 | rs107163178-up | TTCTCCCACCGGAGACTTAG |
| rs107163178-low | GAGAGGGCATATGTGGGATG | |||
| c5 | rs105170847 | chr5 | rs105170847-up | TGGTTGGATGAATGGAGACA |
| rs105170847-low | CCTCAGGAGCACCAACATTT | |||
| c6 | rs106547562 | chr6 | rs106547562-up | GGTTTTGTCCTCAGCAGGTT |
| rs106547562-low | CTCTTAGGCATCTGGGGAAA | |||
| c7 | rs65502317 | chr7 | rs65502317-up | TGTGCATGCTTACAGTGGCTA |
| rs65502317-low | AGCCAACTGGAGCCACATAG | |||
| c8 | rs105694545 | chr8 | rs105694545-up | TCAGCAGTGCTCAGCAGTTT |
| rs105694545-low | CGTAACATCATTTGGCATCG | |||
| c9 | rs65801920 | chr9 | rs65801920-up | TGGCTCTTGGTCATCATCTG |
| rs65801920-low | TTTTGCAGCCTGTGTGTAGC | |||
| c10 | rs105168947 | chr10 | rs105168947-up | GGCGGACAGAACATAGGTGT |
| rs105168947-low | CAGCCAAACTCCCAGATCAT | |||
| c11 | rs106880606 | chr11 | rs106880606-up | AGTGGGTTTAGCAAGCAGGA |
| rs106880606-low | GTCGTCGAAATGCAGGAGAT | |||
| d1 | rs8158863 | chr12 | rs8158863-up | GGTCACAGCCATTCTGGATT |
| rs8158863-low | GCCCTGAATGCAGGTGTAGT | |||
| d2 | rs105965886 | chr13 | rs105965886-up | CACCAGCAAAAGGGAAAAAG |
| rs105965886-low | CAGAACACAGCCTCAGTTGG | |||
| d3 | rs63919332 | chr14 | rs63919332-up | GTCCTGTCGCCCAATATCAC |
| rs63919332-low | ACAACAATGGCAGTGGTCAA | |||
| d4 | rs65809306 | chr15 | rs65809306-up | AGCACTTGTGTGGGAAGCTC |
| rs65809306-low | CCTGACAAAACCACATGCAG | |||
| d5 | rs65087257 | chr16 | rs65087257-up | CCTTCCTTTTCCACTGTCCA |
| rs65087257-low | CTTAGGGCCTAGGGGCTTTA | |||
| d6 | rs106021355 | chr17 | rs106021355-up | GGTGGTTTATGGCCAGGATA |
| rs106021355-low | AACTGGAGAGGAAAAGGGACA | |||
| d7 | rs106171441 | chr18 | rs106171441-up | TCAATTCTTGGCAGCATCAG |
| rs106171441-low | CTGGCCACTTCTCTGAGTCC | |||
| d8 | rs106777713 | chr19 | rs106777713-up | GTGAAGCGACATCGTCAAGA |
| rs106777713-low | CTTGGCTGTGAGATCGAGGT | |||
| d9 | rs104937988 | chr20 | rs104937988-up | GCTGCACAGAACTGATTCCA |
| rs104937988-low | TGCAAGAATTAAATAAAATCCAAA | |||
| d10 | rs105294203 | chrX | rs105294203-up | ACTCCCCAGAGGATTGTCG |
| rs105294203-low | ATGTTCAAGTGCAACGATGG |
表1 40个SNP位点及引物序列
Table 1 40 SNP loci and primer sequences
序号 No. | SNP位点 Reference SNP ID | 染色体 Chromosome | 引物名称 Primer name | 引物序列 Primer sequences |
|---|---|---|---|---|
| a1 | rs65970293 | chr1 | rs65970293-up | TGCACATTGGGACTGCTTTA |
| rs65970293-low | ATCTGGCATGGACAGAGCTT | |||
| a2 | rs106996961 | chr2 | rs106996961-up | ATAGGGCCAAGGTCCCATTC |
| rs106996961-low | GAAAAGAACGCTAATCACAGGACA | |||
| a3 | rs63860341 | chr3 | rs63860341-up | CAATGAATCAAAGGCAACCA |
| rs63860341-low | CCACGGTCCCTATGTGAACT | |||
| a4 | rs64750237 | chr4 | rs64750237-up | CAGGTATGGCTTTGGAGGAA |
| rs64750237-low | GTTTTCAGCTGGAGCTGTCC | |||
| a5 | rs107261116 | chr5 | rs107261116-up | CAGCATTGGCAAGTCTTCCT |
| rs107261116-low | TGATAAAAGGAGCAGCAGCA | |||
| a6 | rs64102787 | chr6 | rs64102787-up | CGGGGATCAAAGAGACAAAA |
| rs64102787-low | ATAGTGCCAAAGCCCAACAC | |||
| a7 | rs105187832 | chr7 | rs105187832-up | AGACTACTGGGCTGCTCACC |
| rs105187832-low | CATGGCACACCTTTCTTTCA | |||
| a8 | rs64682441 | chr9 | rs64682441-up | GCTGGAAGTCTCTGGAGTGG |
| rs64682441-low | ACAAGTGGACTGGCTCAGGT | |||
| a9 | rs65067166 | chr10 | rs65067166-up | ATCCCTCTCTTTGCCAGGAT |
| rs65067166-low | GTAGGGTGAGGCTGCTTCTG | |||
| a10 | rs8167985 | chr11 | rs8167985-up | ACCCCCTCTTGCTTTCTGTT |
| rs8167985-low | CTGCTCTTGGATGAAAACCA | |||
| b1 | rs106672258 | chr12 | rs106672258-up | GCCCTTCCCAAGGTCATACT |
| rs106672258-low | AAAGCACAAAAATGCCATCC | |||
| b2 | rs65921305 | chr13 | rs65921305-up | AACCTGGTGATTTGTCAGAGC |
| rs65921305-low | GGCAGACTCCAGGAAGATTG | |||
| b3 | rs106458396 | chr15 | rs106458396-up | AGAGCAGCTGCCAAAATAGC |
| rs106458396-low | AAACCTGGCTTTAGCAAGCA | |||
| b4 | rs106888236 | chr16 | rs106888236-up | TGTGAAGAACACAGGCTCTCC |
| rs106888236-low | TGCCAAACACCATGTAATGAA | |||
| b5 | rs63814610 | chr17 | rs63814610-up | TAGGACAGGGGGAACAACTG |
| rs63814610-low | CCCAAGCATCCTTGACTGTT | |||
| b6 | rs8152541 | chr18 | rs8152541-up | CGTGTCATTCCTGACTGTGG |
| rs8152541-low | ACAGGAGCTGGCACTGAATC | |||
| b7 | rs105824121 | chr19 | rs105824121-up | TTCTGCAATCAAGGGTGGAT |
| rs105824121-low | GGCCACCAAAGATACACAGG | |||
| b8 | rs13453252 | chr20 | rs13453252-up | TACGCCATTTTGCGACATTA |
| rs13453252-low | GGTTCCCTACGAGGAACGAC | |||
| b9 | rs65108991 | chrX | rs65108991-up | CATCCCAGAAGCCCTGTTTA |
| rs65108991-low | GAAGTCAGTGCTGTGCCTCA | |||
| c1 | rs106891788 | chr1 | rs106891788-up | TCCATCTGCTTGGCCCTAAT |
| rs106891788-low | GTTGCAACCCACAGGTTGAG | |||
| c2 | rs107493604 | chr2 | rs107493604-up | TCCTCCTGGAAGACAGCAAC |
| rs107493604-low | GTTCCTGAGAAGGGGCTCTC | |||
| c3 | rs65632519 | chr3 | rs65632519-up | CCCTGTTTCAGCCTTTTGTG |
| rs65632519-low | GAGGAAGAGGGAAGGGAGAA | |||
| c4 | rs107163178 | chr4 | rs107163178-up | TTCTCCCACCGGAGACTTAG |
| rs107163178-low | GAGAGGGCATATGTGGGATG | |||
| c5 | rs105170847 | chr5 | rs105170847-up | TGGTTGGATGAATGGAGACA |
| rs105170847-low | CCTCAGGAGCACCAACATTT | |||
| c6 | rs106547562 | chr6 | rs106547562-up | GGTTTTGTCCTCAGCAGGTT |
| rs106547562-low | CTCTTAGGCATCTGGGGAAA | |||
| c7 | rs65502317 | chr7 | rs65502317-up | TGTGCATGCTTACAGTGGCTA |
| rs65502317-low | AGCCAACTGGAGCCACATAG | |||
| c8 | rs105694545 | chr8 | rs105694545-up | TCAGCAGTGCTCAGCAGTTT |
| rs105694545-low | CGTAACATCATTTGGCATCG | |||
| c9 | rs65801920 | chr9 | rs65801920-up | TGGCTCTTGGTCATCATCTG |
| rs65801920-low | TTTTGCAGCCTGTGTGTAGC | |||
| c10 | rs105168947 | chr10 | rs105168947-up | GGCGGACAGAACATAGGTGT |
| rs105168947-low | CAGCCAAACTCCCAGATCAT | |||
| c11 | rs106880606 | chr11 | rs106880606-up | AGTGGGTTTAGCAAGCAGGA |
| rs106880606-low | GTCGTCGAAATGCAGGAGAT | |||
| d1 | rs8158863 | chr12 | rs8158863-up | GGTCACAGCCATTCTGGATT |
| rs8158863-low | GCCCTGAATGCAGGTGTAGT | |||
| d2 | rs105965886 | chr13 | rs105965886-up | CACCAGCAAAAGGGAAAAAG |
| rs105965886-low | CAGAACACAGCCTCAGTTGG | |||
| d3 | rs63919332 | chr14 | rs63919332-up | GTCCTGTCGCCCAATATCAC |
| rs63919332-low | ACAACAATGGCAGTGGTCAA | |||
| d4 | rs65809306 | chr15 | rs65809306-up | AGCACTTGTGTGGGAAGCTC |
| rs65809306-low | CCTGACAAAACCACATGCAG | |||
| d5 | rs65087257 | chr16 | rs65087257-up | CCTTCCTTTTCCACTGTCCA |
| rs65087257-low | CTTAGGGCCTAGGGGCTTTA | |||
| d6 | rs106021355 | chr17 | rs106021355-up | GGTGGTTTATGGCCAGGATA |
| rs106021355-low | AACTGGAGAGGAAAAGGGACA | |||
| d7 | rs106171441 | chr18 | rs106171441-up | TCAATTCTTGGCAGCATCAG |
| rs106171441-low | CTGGCCACTTCTCTGAGTCC | |||
| d8 | rs106777713 | chr19 | rs106777713-up | GTGAAGCGACATCGTCAAGA |
| rs106777713-low | CTTGGCTGTGAGATCGAGGT | |||
| d9 | rs104937988 | chr20 | rs104937988-up | GCTGCACAGAACTGATTCCA |
| rs104937988-low | TGCAAGAATTAAATAAAATCCAAA | |||
| d10 | rs105294203 | chrX | rs105294203-up | ACTCCCCAGAGGATTGTCG |
| rs105294203-low | ATGTTCAAGTGCAACGATGG |
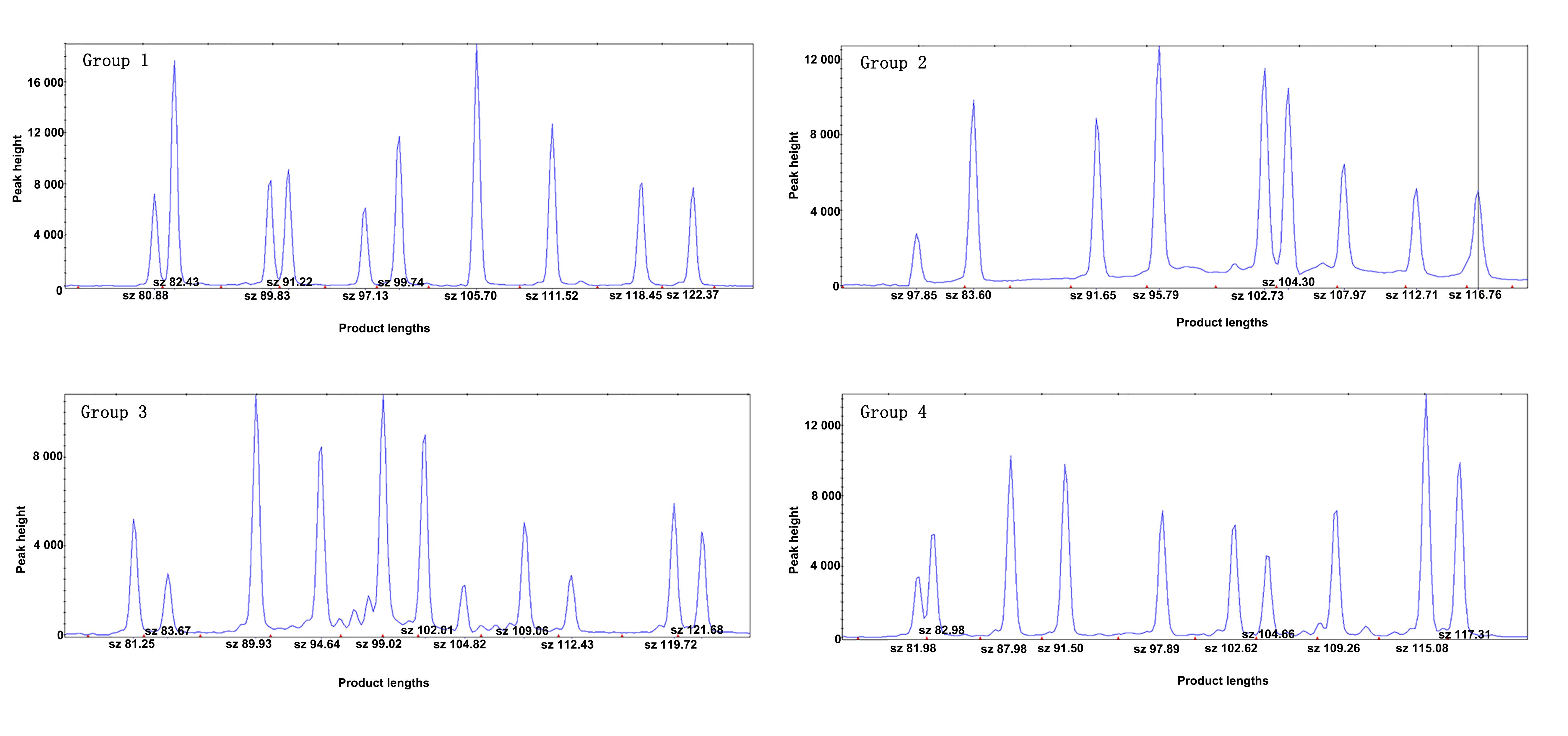
图1 SNP检测方案构建结果(以Lewis/BKl大鼠DNA为模板,采用40个SNP位点进行检测的毛细管电泳峰图)
Figure 1 Results of SNP detection protocol (capillary electrophoresis maps of 40 SNP sites for detection, using the DNA of Lewis/BKl rats as template)
大鼠品系 Rat strains | 碱基有差异的SNP位点编号(右上三角)及数量(左下三角) Differences in SNP loci (right-upper triangle) and numbers (left-lower triangle) | ||||
|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | |
| F344/BKl | - | a1/a2/a3/a4/a5/a9/a10/b2/b5/b7/c2/c3/c6/c8/c10/c11/d2/d6/d7/d8/d9 | a1/a2/a4/a7/a9/b4/b8/c1/c2/c3/c6/c9/c11/d2/d7/d8/d10 | a4/b1/b3/b4/b5/b6/b7/c1/c6/c8/c9/d1/d2 | a2/a4/a7/b2/b7/c1/c2/c4/c5/c7/c8/c11/d1/d2/d7/d8 |
| BN/BKl | 21 | - | a3/a5/a7/a10/b2/b4/b5/b7/b8/c1/c8/c9/c10/d6/d9/d10 | a1/a2/a3/a5/a9/a10/b1/b2/b3/b4/b6/c1/c2/c3/c9/c10/c11/d1/d6/d7/d8/d9 | a1/a3/a5/a7/a9/a10/b5/c1/c3/c4/c5/c6/c7/c10/d1/d6/d9 |
| DA/BKl | 17 | 16 | - | a1/a2/a7/a9/b1/b3/b5/b6/b7/b8/c2/c3/c8/c11/d1/d7/d8/d10 | a1/a9/b2/b4/b7/b8/c3/c4/c5/c6/c7/c8/c9/d1/d10 |
| PVG/BKl | 13 | 22 | 18 | - | a2/a7/b1/b2/b3/b4/b5/b6/c2/c4/c5/c6/c7/c9/c11/d7/d8 |
| Lewis/BKl | 16 | 17 | 15 | 17 | - |
表2 单位A的5个大鼠品系间碱基有差异的SNP位点编号及数量
Table 2 The number and quantity of the SNP loci with different bases among five rat strains from Lab A
大鼠品系 Rat strains | 碱基有差异的SNP位点编号(右上三角)及数量(左下三角) Differences in SNP loci (right-upper triangle) and numbers (left-lower triangle) | ||||
|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | |
| F344/BKl | - | a1/a2/a3/a4/a5/a9/a10/b2/b5/b7/c2/c3/c6/c8/c10/c11/d2/d6/d7/d8/d9 | a1/a2/a4/a7/a9/b4/b8/c1/c2/c3/c6/c9/c11/d2/d7/d8/d10 | a4/b1/b3/b4/b5/b6/b7/c1/c6/c8/c9/d1/d2 | a2/a4/a7/b2/b7/c1/c2/c4/c5/c7/c8/c11/d1/d2/d7/d8 |
| BN/BKl | 21 | - | a3/a5/a7/a10/b2/b4/b5/b7/b8/c1/c8/c9/c10/d6/d9/d10 | a1/a2/a3/a5/a9/a10/b1/b2/b3/b4/b6/c1/c2/c3/c9/c10/c11/d1/d6/d7/d8/d9 | a1/a3/a5/a7/a9/a10/b5/c1/c3/c4/c5/c6/c7/c10/d1/d6/d9 |
| DA/BKl | 17 | 16 | - | a1/a2/a7/a9/b1/b3/b5/b6/b7/b8/c2/c3/c8/c11/d1/d7/d8/d10 | a1/a9/b2/b4/b7/b8/c3/c4/c5/c6/c7/c8/c9/d1/d10 |
| PVG/BKl | 13 | 22 | 18 | - | a2/a7/b1/b2/b3/b4/b5/b6/c2/c4/c5/c6/c7/c9/c11/d7/d8 |
| Lewis/BKl | 16 | 17 | 15 | 17 | - |
序号 No. | SNP标记名称 Reference SNP ID | 染色体 Chromosome | 不同品系大鼠的基因型 Genotype of different strains of rats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | SHR/Slac | GK/Slac | F344/JclSlac | Wistar/Slac | WKY/Slac | CXB | CHN1 | CHN2 | CHN5 | |||
| a1 | rs65970293 | chr1 | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | T/T | T/T | T/T | T/T | T/T |
| a2 | rs106996961 | chr2 | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | T/T |
| a3 | rs63860341 | chr3 | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| a4 | rs64750237 | chr4 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| a5 | rs107261116 | chr5 | C/C | G/G | C/C | C/C | C/C | G/G | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G |
| a6 | rs64102787 | chr6 | C/C | C/C | C/C | C/C | C/C | A/A | A/A | C/C | C/C | A/A | C/C | C/C | C/C | A/A |
| a7 | rs105187832 | chr7 | G/G | G/G | T/T | G/G | T/T | G/G | T/T | G/G | T/T | T/T | G/G | G/G | G/G | G/G |
| a8 | rs64682441 | chr9 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| a9 | rs65067166 | chr10 | C/C | G/G | G/G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G | G/G | G/G |
| a10 | rs8167985 | chr11 | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| b1 | rs106672258 | chr12 | T/T | T/T | T/T | C/C | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| b2 | rs65921305 | chr13 | G/G | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | T/T |
| b3 | rs106458396 | chr15 | T/T | T/T | T/T | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| b4 | rs106888236 | chr16 | C/C | C/C | A/A | A/A | C/C | A/A | A/A | C/C | A/A | C/C | C/C | A/A | A/A | C/C |
| b5 | rs63814610 | chr17 | T/T | C/C | T/T | C/C | T/T | T/T | T/T | T/T | C/C | T/T | C | T/T | T/T | T/T |
| b6 | rs8152541 | chr18 | G/G | G/G | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | A/A | A/A | G/G | A/A |
| b7 | rs105824121 | chr19 | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | G/G | G/G | A/A | A/A | A/A | G/G |
| b8 | rs13453252 | chr20 | G/G | G/G | C/C | G/G | G/G | G/G | C/C | G/G | C/C | C/C | G/G | G/G | G/G | G/G |
| b9 | rs65108991 | chrX | A/A | A/A | A/A | A/A | A/A | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A |
| c1 | rs106891788 | chr1 | C/C | C/C | T/T | T/T | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| c2 | rs107493604 | chr2 | A/A | G/G | G/G | A/A | G/G | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | G/G |
| c3 | rs65632519 | chr3 | T/T | A/A | A/A | T/T | T/T | A/A | A/A | T/T | T/T | T/T | A | T/T | T/T | T/T |
| c4 | rs17163178 | chr4 | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | T/T | T/T | C/C |
| c5 | rs105170847 | chr5 | C/C | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| c6 | rs106547562 | chr6 | A/A | C/C | C/C | C/C | A/A | C/C | C/C | A/A | A/A | C/C | A/A | A/A | A/A | C/C |
| c7 | rs65502317 | chr7 | C/C | C/C | C/C | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A | A/A | C/C |
| c8 | rs105694545 | chr8 | G/G | A/A | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | G/G | A/A | A/A |
| c9 | rs65801920 | chr9 | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| c10 | rs105168947 | chr10 | C/C | T/T | C/C | C/C | C/C | T/T | T/T | C/C | C/C | C/C | T/T | T/T | T/T | C/C |
| c11 | rs106880606 | chr11 | G/G | T/T | T/T | G/G | T/T | T/T | T/T | G/G | T/T | T/T | T/T | T/T | T/T | G/G |
| d1 | rs8158863 | chr12 | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | A/A | G/G | A/A | A/A | A/A | G/G |
| d2 | rs105965886 | chr13 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C |
| d3 | rs63919332 | chr14 | C/C | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | C/C |
| d4 | rs65809306 | chr15 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | T/T | T/T | T/T | C/C |
| d5 | rs65087257 | chr16 | A/A* | A/A | A/A | A/A | A/A | A/A | C/C | C/C* | C/C | A/A | C/C | A/A | A/A | C/C |
| d6 | rs106021355 | chr17 | T/T | C/C | T/T | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| d7 | rs106171441 | chr18 | G/G | A/A | A/A | G/G | A/A | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A | A/A |
| d8 | rs106777713 | chr19 | G/G* | A/A | A/A | G/G | A/A | T/T | C/C | T/T* | T/T | C/C | C/C | T/T | T/T | T/T |
| d9 | rs104937988 | chr20 | T/T | G/G | T/T | T/T | T/T | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | G/G |
| d10 | rs105294203 | chrX | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | G/G | G/G | G/G |
表3 国内不同供应商来源的常用近交系大鼠的SNP多重PCR-LDR检测结果 (supplies)
Table 3 Multiplex PCR-ligase detection reaction (LDR) test results of inbred rat strains from different domestic animal
序号 No. | SNP标记名称 Reference SNP ID | 染色体 Chromosome | 不同品系大鼠的基因型 Genotype of different strains of rats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | SHR/Slac | GK/Slac | F344/JclSlac | Wistar/Slac | WKY/Slac | CXB | CHN1 | CHN2 | CHN5 | |||
| a1 | rs65970293 | chr1 | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | T/T | T/T | T/T | T/T | T/T |
| a2 | rs106996961 | chr2 | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | T/T |
| a3 | rs63860341 | chr3 | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| a4 | rs64750237 | chr4 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| a5 | rs107261116 | chr5 | C/C | G/G | C/C | C/C | C/C | G/G | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G |
| a6 | rs64102787 | chr6 | C/C | C/C | C/C | C/C | C/C | A/A | A/A | C/C | C/C | A/A | C/C | C/C | C/C | A/A |
| a7 | rs105187832 | chr7 | G/G | G/G | T/T | G/G | T/T | G/G | T/T | G/G | T/T | T/T | G/G | G/G | G/G | G/G |
| a8 | rs64682441 | chr9 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| a9 | rs65067166 | chr10 | C/C | G/G | G/G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G | G/G | G/G |
| a10 | rs8167985 | chr11 | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| b1 | rs106672258 | chr12 | T/T | T/T | T/T | C/C | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| b2 | rs65921305 | chr13 | G/G | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | T/T |
| b3 | rs106458396 | chr15 | T/T | T/T | T/T | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| b4 | rs106888236 | chr16 | C/C | C/C | A/A | A/A | C/C | A/A | A/A | C/C | A/A | C/C | C/C | A/A | A/A | C/C |
| b5 | rs63814610 | chr17 | T/T | C/C | T/T | C/C | T/T | T/T | T/T | T/T | C/C | T/T | C | T/T | T/T | T/T |
| b6 | rs8152541 | chr18 | G/G | G/G | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | A/A | A/A | G/G | A/A |
| b7 | rs105824121 | chr19 | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | G/G | G/G | A/A | A/A | A/A | G/G |
| b8 | rs13453252 | chr20 | G/G | G/G | C/C | G/G | G/G | G/G | C/C | G/G | C/C | C/C | G/G | G/G | G/G | G/G |
| b9 | rs65108991 | chrX | A/A | A/A | A/A | A/A | A/A | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A |
| c1 | rs106891788 | chr1 | C/C | C/C | T/T | T/T | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| c2 | rs107493604 | chr2 | A/A | G/G | G/G | A/A | G/G | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | G/G |
| c3 | rs65632519 | chr3 | T/T | A/A | A/A | T/T | T/T | A/A | A/A | T/T | T/T | T/T | A | T/T | T/T | T/T |
| c4 | rs17163178 | chr4 | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | T/T | T/T | C/C |
| c5 | rs105170847 | chr5 | C/C | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| c6 | rs106547562 | chr6 | A/A | C/C | C/C | C/C | A/A | C/C | C/C | A/A | A/A | C/C | A/A | A/A | A/A | C/C |
| c7 | rs65502317 | chr7 | C/C | C/C | C/C | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A | A/A | C/C |
| c8 | rs105694545 | chr8 | G/G | A/A | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | G/G | A/A | A/A |
| c9 | rs65801920 | chr9 | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| c10 | rs105168947 | chr10 | C/C | T/T | C/C | C/C | C/C | T/T | T/T | C/C | C/C | C/C | T/T | T/T | T/T | C/C |
| c11 | rs106880606 | chr11 | G/G | T/T | T/T | G/G | T/T | T/T | T/T | G/G | T/T | T/T | T/T | T/T | T/T | G/G |
| d1 | rs8158863 | chr12 | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | A/A | G/G | A/A | A/A | A/A | G/G |
| d2 | rs105965886 | chr13 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C |
| d3 | rs63919332 | chr14 | C/C | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | C/C |
| d4 | rs65809306 | chr15 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | T/T | T/T | T/T | C/C |
| d5 | rs65087257 | chr16 | A/A* | A/A | A/A | A/A | A/A | A/A | C/C | C/C* | C/C | A/A | C/C | A/A | A/A | C/C |
| d6 | rs106021355 | chr17 | T/T | C/C | T/T | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| d7 | rs106171441 | chr18 | G/G | A/A | A/A | G/G | A/A | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A | A/A |
| d8 | rs106777713 | chr19 | G/G* | A/A | A/A | G/G | A/A | T/T | C/C | T/T* | T/T | C/C | C/C | T/T | T/T | T/T |
| d9 | rs104937988 | chr20 | T/T | G/G | T/T | T/T | T/T | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | G/G |
| d10 | rs105294203 | chrX | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | G/G | G/G | G/G |
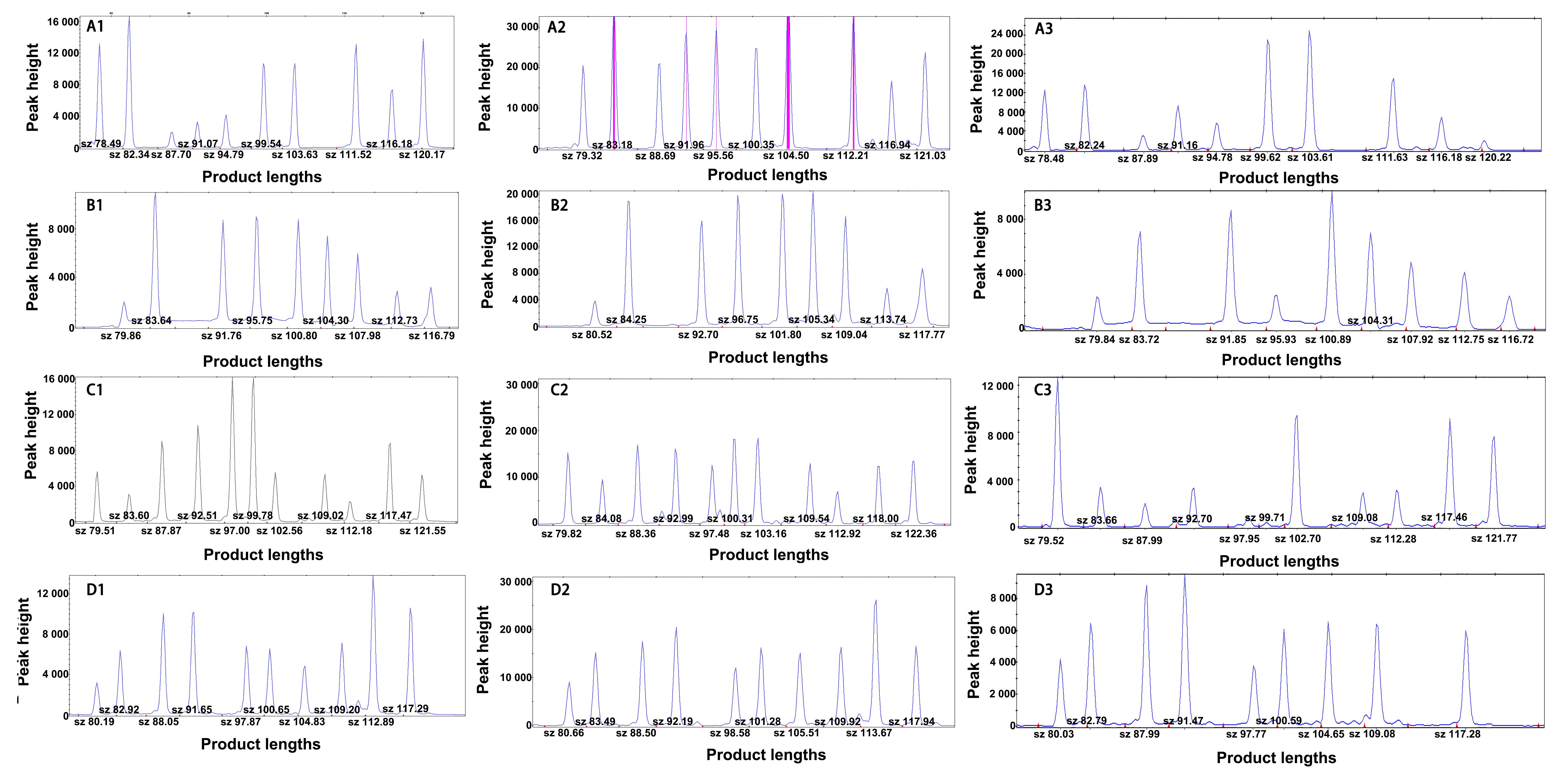
图2 大鼠SNP多重PCR-LDR检测方案中3种DNA聚合酶的应用结果(以BN/BKl大鼠为例)注:A1~A3为用酶1、酶2、酶3检测的第1组SNP位点结果;B1~B3为酶1、酶2、酶3检测的第2组SNP位点结果;C1~C3为酶1、酶2、酶3检测的第3组SNP位点结果;D1~D3为酶1、酶2、酶3检测的第4组SNP位点结果。
Figure 2 Application results of three DNA polymerases in the multiple PCR-LDR detection scheme of rat SNP (using BN/BKl rats as an example)Note: A1-A3, Results of the first group of SNP loci detected using enzyme 1, enzyme 2, and enzyme 3, respectively; B1-B3, Results of the second group of SNP loci detected for enzyme 1, enzyme 2, and enzyme 3; C1-C3, Results of the third group of SNP loci detected by enzyme 1, enzyme 2, and enzyme 3; D1-D3, Results of the fourth group of SNP loci detected for enzyme 1, enzyme 2, and enzyme 3.
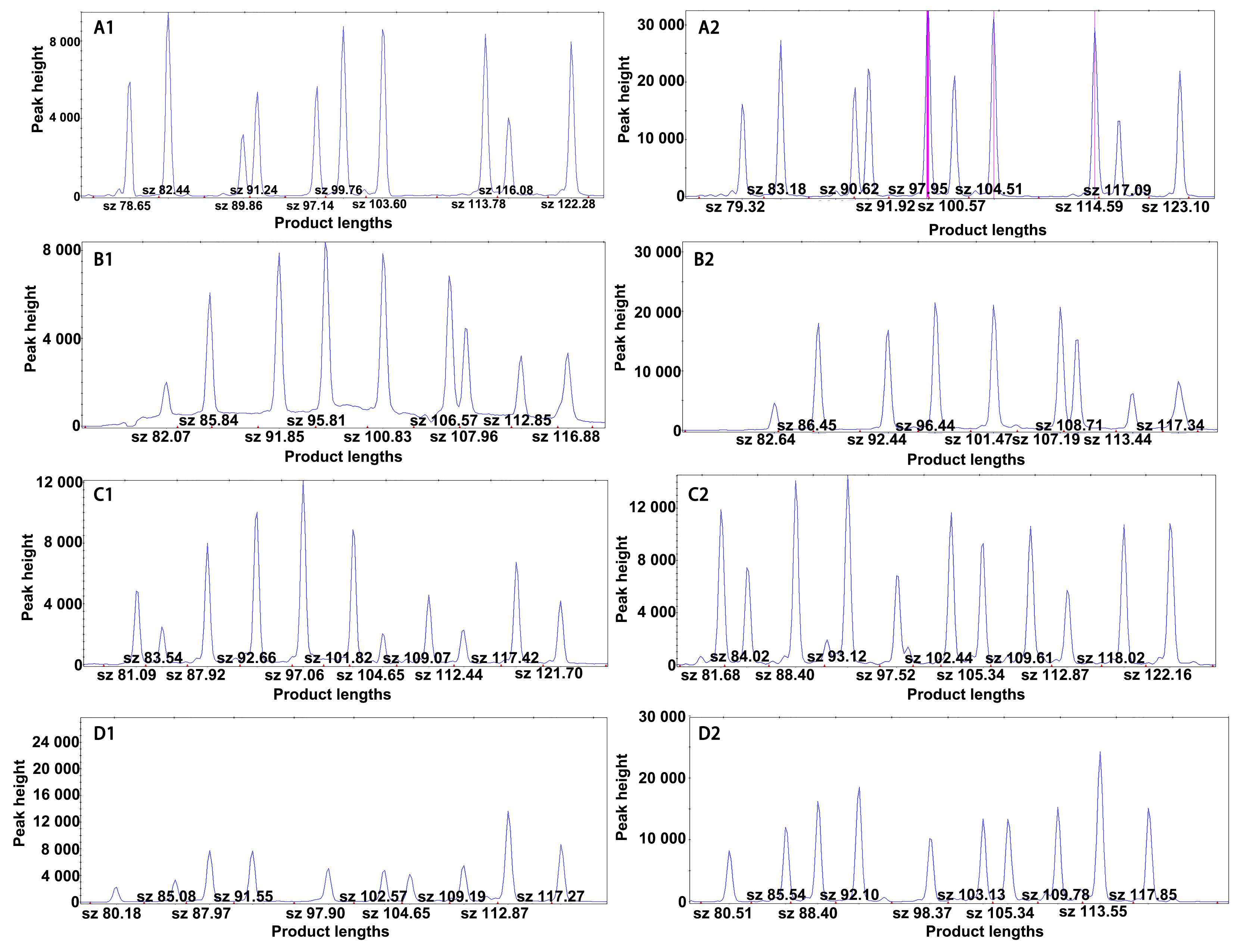
图3 不同实验室间大鼠SNP多重PCR-LDR检测结果比较(以CXB大鼠为例)注:A1、B1、C1、D1分别为本机构检测的第1组、第2组、第3组、第4组SNP位点的电泳结果峰图;A2、B2、C2、D2分别为第三方实验室检测的第1组、第2组、第3组、第4组SNP位点的电泳结果峰图。
Figure 3 Comparison of multiplex PCR-ligase detection reaction (LDR) results among different laboratories (using CXB rats as an example)Note: A1-D1, electrophoretic peak maps of SNP sites in group 1- 4 tested by our institution. A2-D2, electrophoretic peak maps of SNP sites in group 1- 4 tested by the third-party laboratory.
| 1 | BENAVIDES F, RÜLICKE T, PRINS J B, et al. Genetic quality assurance and genetic monitoring of laboratory mice and rats: FELASA Working Group Report[J]. Lab Anim, 2020, 54(2):135-148. DOI: 10.1177/0023677219867719 . |
| 2 | HOFFMAN H A. 实验动物遗传质量的控制重点在遗传监测[J]. 上海实验动物科学, 1987, 7(1): 55-57. |
| HOFFMAN H A. The key to controlling the genetic quality of experimental animals is genetic monitoring[J]. Shanghai Lab Anim Sci, 1987, 7(1): 55-57. | |
| 3 | 国家市场监督管理总局, 国家标准化管理委员会. 实验动物 遗传质量控制: GB 14923—2022[S]. 北京: 中国标准出版社, 2022. |
| State Administration for Market Regulation, Standardization Administration of the People's Republic of China. Laboratory animal-Genetic quality control: GB 14923-2022[S]. Beijing: Standards Press of China, 2022 | |
| 4 | 杨玲焰,王立鹏,时长军. 一种基于KASP的大鼠遗传质量监控SNP标记分型方法及试剂盒: CN201910648812X[P/OL]. 2019-11-15[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| YANG L Y, WANG L P, SHI C J. A SNP marker typing method and kit based KASP for monitoring rat genetic quality: CN201910648812X[P/OL]. 2019-11-15[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 5 | 琚存祥, 赵静, 马秀英, 等. 一种近交系遗传质量监控的SNP快速检测方法和SNP位点及其引物: CN108588236A[P/OL]. 2018-09-28[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| JU C X, ZHAO J, MA X Y, et al. Rapid SNP detection method for genetic quality monitoring of inbred lines and SNP sites and primers: CN108588236A[P/OL]. 2018-09-28[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 6 | 琚存祥, 赵静, 杨旭乐, 等. 一组用于BALB/cJ近交系小鼠遗传质量监控的SNP位点及其引物组合和应用: CN110358847A[P/OL]. 2019-10-22[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| JU C X, ZHAO J, YANG X L, et al. A set of SNP sites and primer combinations and applications for genetic quality control in BALB/cJ inbred mice: CN110358847A[P/OL]. 2019-10-22[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 7 | 赵静, 琚存祥, 马秀英, 等. 一组用于CBA/CaJ近交系小鼠遗传质量监控的SNP位点及其引物组合和应用: CN109609659A[P/OL]. 2019-04-12[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| ZHAO J, JU C X, MA X Y, et al. A set of SNP sites and their primer combinations and applications for genetic quality monitoring in CBA/CaJ inbred mice: CN109609659A[P/OL]. 2019-04-12[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 8 | 王刚, 李凯, 周宇荀, 等. 新型通用探针LDR分型技术的开发及细胞色素P450基因多位点分型[J]. 华东理工大学学报(自然科学版), 2008, 34(4):503-508. DOI: 10.3969/j.issn.1006-3080.2006.09.008 . |
| WANG G, LI K, ZHOU Y X, et al. Improvement of novel LDR genotyping system based on universal probe and the SNPs typing of human P450[J]. J East China Univ Sci Technol Nat Sci Ed, 2008, 34(4):503-508. DOI: 10.3969/j.issn.1006-3080.2006.09.008 . | |
| 9 | 徐园, 钱强, 鲍世民, 等. 基于高通量靶向测序的大鼠遗传质量检测方案建立[J]. 生物化工, 2020, 6(4): 65-69. DOI: 10.3969/j.issn.2096-0387.2020.04.016 . |
| XU Y, QIAN Q, BAO S M, et al. Establishment of rat genetic quality detection scheme based on high throughput targeted sequencing[J]. Biol Chem Eng, 2020, 6(4): 65-69. DOI: 10.3969/j.issn.2096-0387.2020.04.016 . | |
| 10 | SAAR K, BECK A, BIHOREAU M T, et al. SNP and haplotype mapping for genetic analysis in the rat[J]. Nat Genet, 2008, 40(5):560-566. DOI: 10.1038/ng.124 . |
| 11 | PETKOV P M, DING Y M, CASSELL M A, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships[J]. Genome Res, 2004, 14(9):1806-1811. DOI: 10.1101/gr.2825804 . |
| 12 | TAFT R A, DAVISSON M, WILES M V. Know thy mouse[J]. Trends Genet, 2006, 22(12):649-653. DOI: 10.1016/j.tig.2006.09.010 . |
| 13 | MEKADA K, HIROSE M, MURAKAMI A, et al. Development of SNP markers for C57BL/6N-derived mouse inbred strains[J]. Exp Anim, 2015, 64(1):91-100. DOI: 10.1538/expanim.14-0061 . |
| [1] | 刘巍, 张心妍, 侯丰田, 许中衎, 马丽颖. 实验动物设施换气次数检测能力验证结果评价[J]. 实验动物与比较医学, 2025, 45(1): 87-95. |
| [2] | 乔涵, 魏杰, 王菲菲, 王洪, 邢进, 付瑞, 肖镜, 赵萌, 岳秉飞, 项新华. 2013—2022年实验动物检测能力验证样品的回顾性研究[J]. 实验动物与比较医学, 2022, 42(6): 483-489. |
| [3] | 李晓波, 王吉, 王洪, 王淑菁, 王莎莎, 秦骁, 李威, 岳秉飞, 付瑞. 猪细小病毒核酸检测实验室能力验证结果分析[J]. 实验动物与比较医学, 2022, 42(6): 490-497. |
| [4] | 冯育芳, 王洪, 付瑞, 张雪青, 高强, 岳秉飞, 邢进. 实验动物四种呼吸道病原菌实验室检测能力验证结果分析[J]. 实验动物与比较医学, 2022, 42(6): 498-504. |
| [5] | 魏杰, 张心妍, 王洪, 赵蓝, 刘巍, 李欢, 付瑞, 乔涵, 赵萌, 项新华, 岳秉飞. 实验小鼠核酸样品单核苷酸多态性标记检测的能力验证结果评价[J]. 实验动物与比较医学, 2022, 42(6): 505-510. |
| [6] | 郑杰, 张国忠, 胡艳梅, 伍学一, 杨宗富, 字向东. 基于RNA-Seq技术的犏牛囊胚冷冻前后单核苷酸多态性和可变剪切分析[J]. 实验动物与比较医学, 2020, 40(4): 279-. |
| [7] | 钱强, 徐园, 王亚恒, 周宇荀, 肖君华, 韩琳, 鲍世民, 李凯. 基于多重PCR靶向二代测序的近交系小鼠遗传质量监测方法建立[J]. 实验动物与比较医学, 2019, 39(2): 111-117. |
| [8] | 高一龙, 贺星亮, 强京宁, 宋珍华, 温海, 陶晓冉, 孙勇, 秦海斌, 包喜军, 孟戈, 朱骞. 基于Sequenom MassARRAY SNP技术检测嗅觉受体基因与工作犬嗅探能力的关联性研究[J]. 实验动物与比较医学, 2019, 39(2): 140-145. |
| [9] | 李银银, 陈振文, 刘丽均, 李长龙, 刘欣, 吕建祎, 郭萌, 杜小燕. 单核苷酸多态性位点法对国内近交系小鼠遗传检测[J]. 实验动物与比较医学, 2018, 38(1): 16-21. |
| [10] | 韩琳, 谢建云, 杨斐, 胡樱, 田立立, 鲍世民. 上海地区常用近交系小鼠品系的单核苷酸多态性分型研究[J]. 实验动物与比较医学, 2017, 37(1): 25-31. |
| [11] | 赵丽亚, 张蓉, 赵莹, 邢正弘, 陈国强. 单核苷酸多态性(SNP)在近交系小鼠遗传检测中的应用[J]. 实验动物与比较医学, 2016, 36(1): 24-31. |
| [12] | 贺争鸣, 岳秉飞, 巩薇, 付瑞, 邢进, 王洪. 加强实验动物质量检测实验室参与能力验证工作[J]. 实验动物与比较医学, 2014, 34(5): 393-395. |
| [13] | 林丽芳, 王洪, 赵善民, 肖邦, 孙智远, 张璐, 袁子彦, 于琛琳, 蔡丽萍, 徐晨, 汤球, 孙伟, 崔淑芳. 裸鼹鼠生化位点的初步研究[J]. 实验动物与比较医学, 2013, 33(6): 477-480. |
| [14] | 姜宪环, 蔡慧强, 高骏, 倪丽菊, 李凯, 肖君华, 周宇荀. 用LA-PCR方法克隆东方田鼠部分Y染色体序列[J]. 实验动物与比较医学, 2012, 32(4): 324-328. |
| [15] | 程树军, 秦瑶, 步犁. 体外重建人体皮肤模型刺激试验的验证[J]. 实验动物与比较医学, 2012, 32(3): 243-246. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||