













实验动物与比较医学 ›› 2025, Vol. 45 ›› Issue (6): 773-783.DOI: 10.12300/j.issn.1674-5817.2025.138
贠佳琦1,2, 马芹1,2, 王官栋1, 孙佩璐1, 王义冠1, 王四宝1( )(
)( )
)
收稿日期:2025-09-05
修回日期:2025-10-14
出版日期:2025-12-25
发布日期:2025-12-19
作者简介:贠佳琦(1997—),女,博士研究生,研究方向:医学媒介昆虫与微生物互作。E-mail: yunjiaqi@cemps.ac.cn;基金资助:
YUN Jiaqi1,2, MA Qin1,2, WANG Guandong1, SUN Peilu1, WANG Yiguan1, WANG Sibao1( )(
)( )
)
Received:2025-09-05
Revised:2025-10-14
Published:2025-12-25
Online:2025-12-19
Contact:
WANG Sibao (ORCID:0000-0002-5880-0815), E-mail: sbwang@cemps.ac.cn摘要:
蚊媒疾病(如疟疾、登革热、寨卡病毒病、基孔肯雅热等)对全球公共卫生造成了重大威胁,然而基于化学农药的传统防控手段面临着媒介蚊虫耐药性增强、污染环境等严峻挑战。遗传控制策略因具有物种特异性及环境友好等优势,成为蚊媒控制中极具潜力的替代方案。基因驱动(gene drive)技术利用成簇规律间隔短回文重复序列(clustered regularly interspaced short palindromic repeats,CRISPR)/CRISPR相关蛋白核酸酶9(CRISPR-associated nuclease 9,Cas9)等基因编辑工具,使特定基因以超孟德尔遗传的方式在目标蚊种群中高效传播,为蚊媒疾病防控提供了革命性策略。本文系统综述了基因驱动技术在该领域的关键进展、核心挑战及应对策略。研究进展:(1)在疟疾媒介按蚊中,靶向性别决定基因或雌性生殖基因的种群抑制型驱动可致雌性不育或性别失衡,实现种群抑制;靶向按蚊中与疟原虫感染相关的宿主基因或递送抗疟疾效应分子的种群替代型基因驱动策略,可有效阻断病原体传播。(2)在蚊媒病毒媒介伊蚊中,靶向雌蚊飞行必需基因实现种群抑制,并探索抗病毒效应系统与驱动元件的偶联;优化的分割型基因驱动策略展现出高切割与重组效率,模型预测可实现抗病性状的安全可控扩散。(3)在淋巴丝虫病媒介库蚊中,将同源驱动元件分别整合至两个参与眼睛色素合成通路的基因中,可以通过眼色观察到明显的基因驱动效率。核心挑战:技术层面存在同源重组修复效率低、非同源末端连接修复导致抗性等位基因产生、CRISPR/Cas9脱靶效应及物种适配性差异;生态与安全层面涉及驱动元件意外扩散导致的基因池污染、生态平衡潜在影响及长期不可逆性风险。应对策略与展望:采用多重向导RNA(guide RNA,gRNA)靶向策略以提升驱动稳定性和对抗潜在抗性;开发可逆性设计,如合成抗性、逆转驱动及免疫性逆转驱动作为“基因刹车”;建立长期生态监测系统与数学模型进行风险评估;探索“环境响应型驱动”以增强可控性。未来研究亟须持续优化驱动效率与特异性,深化生态风险评估,加强跨国合作,并推动伦理共识与监管框架构建,以期在安全可控前提下,使基因驱动技术成为应对蚊媒疾病这一全球健康挑战的可持续性防控策略。
中图分类号:
贠佳琦,马芹,王官栋,等. 基因驱动技术在蚊媒疾病防控中的研究进展与挑战[J]. 实验动物与比较医学, 2025, 45(6): 773-783. DOI: 10.12300/j.issn.1674-5817.2025.138.
YUN Jiaqi,MA Qin,WANG Guandong,et al. Research Advances and Challenges of Gene Drive Technology in Mosquito-Borne Disease Control[J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 773-783. DOI: 10.12300/j.issn.1674-5817.2025.138.
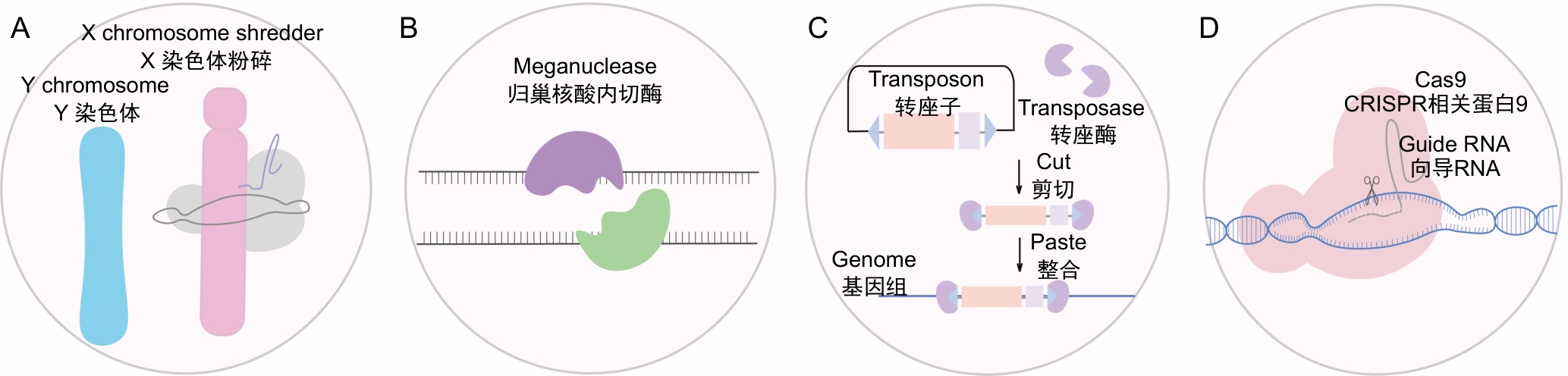
图1 基于不同驱动元件的基因驱动类型注:A,减数分裂介导的基因驱动,X染色体粉碎导致强烈的性别比例扭曲,后代趋于雄性;B,基于归巢核酸内切酶的基因驱动,归巢核酸内切酶能够识别并切割特定DNA序列,触发同源重组修复机制;C,基于转座子和转座酶系统的基因驱动,转座酶催化转座子的插入和移动,将目标基因随机插入宿主基因组中;D,基于CRISPR/Cas9系统的基因驱动,将驱动基因定点插入目标基因位点,并利用Cas9蛋白持续切割野生型等位基因。
Figure 1 Gene drive types based on different driver elementsNote: A, Meiosis-mediated gene drive, X chromosome shattering leads to a strong sex ratio distortion, with offspring tending to be male; B, Gene drive based on meganuclease, meganuclease recognizes and cuts specific DNA sequence, triggering the homologous recombination repair mechanism; C, Gene drive based on transposon and transposase systems, transposases catalyze the insertion and movement of transposons, randomly inserting target genes into the host genome; D, Gene drive based on the CRISPR/Cas9 system, the drive gene is precisely inserted into the target gene locus, and the Cas9 protein continuously cuts the wild-type allele.
图2 基于功能划分的基因驱动类型注:A,种群抑制型基因驱动,通过设计具有“自私遗传”特性的基因元件,在蚊媒种群中快速扩散致死性或生殖干扰性基因,最终实现种群数量抑制;B,种群替代型基因驱动,通过基因编辑赋予蚊媒抗病特性,在不显著改变种群数量的前提下阻断病原体传播链。
Figure 2 Gene drive types based on functional classificationNote: A, Population suppression gene drive, by designing genetic elements with "selfish inheritance" characteristics, rapidly spreads lethal or reproduction-interfering genes in the mosquito population, ultimately achieving population suppression; B, Population replacement gene drive: through gene editing, endows mosquitoes with disease-resistance characteristics, blocking the transmission chain of pathogens without significantly altering the population size.
| 减数分裂介导的基因驱动 | Galizi R, | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Kyrou K, | ||
| Hammond A, et al. (2016)[ | |||
| fibrinogen-related protein 1 | Dong Y, et al. (2018)[ | ||
| cardinal | Carballar-lejarazu R, et al. (2020)[ | ||
| single chain variable fragment | Carballar-lejarazu R, et al. (2023)[ | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Xu X, et al. (2025)[ | ||
| Gantz V M, et al. (2015)[ | |||
| CRISPR/Cas9介导的基因驱动 | Green E I, et al. (2023)[ | ||
致倦库蚊 Culex quinquefasciatus | CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | Feng X C, et al. (2021)[ | |
| CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | O'Leary S, et al. (2020)[ | ||
| Li M, et al. (2020)[ | |||
| 归巢核酸内切酶介导的基因驱动 | Franz A W, et al. (2006)[ Williams A E, et al. (2020)[ | ||
| 转座子介导的基因驱动 | Buchman A, et al. (2020)[ |
表1 基因驱动技术在防控蚊媒疾病中的主要进展
Table 1 Main advances of gene drive technology in mosquito-borne disease control
| 减数分裂介导的基因驱动 | Galizi R, | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Kyrou K, | ||
| Hammond A, et al. (2016)[ | |||
| fibrinogen-related protein 1 | Dong Y, et al. (2018)[ | ||
| cardinal | Carballar-lejarazu R, et al. (2020)[ | ||
| single chain variable fragment | Carballar-lejarazu R, et al. (2023)[ | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Xu X, et al. (2025)[ | ||
| Gantz V M, et al. (2015)[ | |||
| CRISPR/Cas9介导的基因驱动 | Green E I, et al. (2023)[ | ||
致倦库蚊 Culex quinquefasciatus | CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | Feng X C, et al. (2021)[ | |
| CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | O'Leary S, et al. (2020)[ | ||
| Li M, et al. (2020)[ | |||
| 归巢核酸内切酶介导的基因驱动 | Franz A W, et al. (2006)[ Williams A E, et al. (2020)[ | ||
| 转座子介导的基因驱动 | Buchman A, et al. (2020)[ |
| [1] | World Health Organization. Vector-borne diseases[Z/OL]. (2024-09-26)[2025-08-21] . |
| [2] | World Health Organization. Dengue[Z/OL]. (2025-08-21)[2025-08-21]. . |
| [3] | World Health Organization. New WHO guidelines for clinical management of arboviral diseases: dengue, chikungunya, Zika and yellow fever[Z/OL]. (2025-07-10)[2025-08-21]. . |
| [4] | KITTAYAPONG P, NINPHANOMCHAI S, LIMOHPASMANEE W, et al. Combined sterile insect technique and incompatible insect technique: the first proof-of-concept to suppress Aedes aegypti vector populations in semi-rural settings in Thailand[J]. PLoS Negl Trop Dis, 2019, 13(10):e0007771. DOI:10.1371/journal.pntd.0007771 . |
| [5] | HARRIS A F, MCKEMEY A R, NIMMO D, et al. Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes[J]. Nat Biotechnol, 2012, 30(9):828-830. DOI:10.1038/nbt.2350 . |
| [6] | MCMENIMAN C J, LANE R V, CASS B N, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti [J]. Science, 2009, 323(5910):141-144. DOI:10.1126/science.1165326 . |
| [7] | PHUC H K, ANDREASEN M H, BURTON R S, et al. Late-acting dominant lethal genetic systems and mosquito control[J]. BMC Biol, 2007, 5:11. DOI:10.1186/1741-7007-5-11 . |
| [8] | HOFFMANN A A, MONTGOMERY B L, POPOVICI J, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission[J]. Nature, 2011, 476(7361):454-457. DOI:10.1038/nature10356 . |
| [9] | AMUZU H E, TSYGANOV K, KOH C, et al. Wolbachia enhances insect-specific flavivirus infection in Aedes aegypti mosquitoes[J]. Ecol Evol, 2018, 8(11):5441-5454. DOI:10.1002/ece3.4066 . |
| [10] | ZÉLÉ F, NICOT A, BERTHOMIEU A, et al. Wolbachia increases susceptibility to Plasmodium infection in a natural system[J]. Proc Biol Sci, 2014, 281(1779):20132837. DOI:10.1098/rspb.2013.2837 . |
| [11] | BURT A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations[J]. Proc Biol Sci, 2003, 270(1518):921-928. DOI:10.1098/rspb.2002.2319 . |
| [12] | AKBARI O S, BELLEN H J, BIER E, et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory[J]. Science, 2015, 349(6251):927-929. DOI:10.1126/science.aac7932 . |
| [13] | MARSHALL J M, HAY B A. Confinement of gene drive systems to local populations: a comparative analysis[J]. J Theor Biol, 2012, 294:153-171. DOI:10.1016/j.jtbi.2011.10.032 . |
| [14] | CHAMPER J, BUCHMAN A, AKBARI O S. Cheating evolution: engineering gene drives to manipulate the fate of wild populations[J]. Nat Rev Genet, 2016, 17(3):146-159. DOI:10.1038/nrg.2015.34 . |
| [15] | JAMES A A. Gene drive systems in mosquitoes: rules of the road[J]. Trends Parasitol, 2005, 21(2):64-67. DOI:10.1016/j.pt.2004.11.004 . |
| [16] | O'BROCHTA D A, ALFORD R T, PILITT K L, et al. piggyBac transposon remobilization and enhancer detection in Anopheles mosquitoes[J]. Proc Natl Acad Sci USA, 2011, 108(39):16339-16344. DOI:10.1073/pnas.1110628108 . |
| [17] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121):819-823. DOI:10.1126/science.1231143 . |
| [18] | KYROU K, HAMMOND A M, GALIZI R, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes[J]. Nat Biotechnol, 2018, 36(11):1062-1066. DOI:10.1038/nbt.4245 . |
| [19] | HAMMOND A, GALIZI R, KYROU K, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae [J]. Nat Biotechnol, 2016, 34(1):78-83. DOI:10.1038/nbt.3439 . |
| [20] | DONG Y M, SIMÕES M L, MAROIS E, et al. CRISPR/Cas9-mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection[J]. PLoS Pathog, 2018, 14(3):e1006898. DOI:10.1371/journal.ppat.1006898 . |
| [21] | CARBALLAR-LEJARAZÚ R, OGAUGWU C, TUSHAR T, et al. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae [J]. Proc Natl Acad Sci USA, 2020, 117(37):22805-22814. DOI:10.1073/pnas.2010214117 . |
| [22] | GALIZI R, DOYLE L A, MENICHELLI M, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito[J]. Nat Commun, 2014, 5:3977. DOI:10.1038/ncomms4977 . |
| [23] | XU X J, CHEN J H, WANG Y, et al. Gene drive-based population suppression in the malaria vector Anopheles stephensi [J]. Nat Commun, 2025, 16(1):1007. DOI:10.1038/s41467-025-56290-2 . |
| [24] | LI Z Q, DONG Y M, YOU L, et al. Driving a protective allele of the mosquito FREP1 gene to combat malaria[J]. Nature, 2025, 645(8081):746-754. DOI:10.1038/s41586-025-09283-6 . |
| [25] | GREEN E I, JAOUEN E, KLUG D, et al. A population modification gene drive targeting both Saglin and Lipophorin impairs Plasmodium transmission in Anopheles mosquitoes[J]. eLife, 2023, 12:e93142. DOI:10.7554/eLife.93142 . |
| [26] | CARBALLAR-LEJARAZÚ R, DONG Y M, PHAM T B, et al. Dual effector population modification gene-drive strains of the African malaria mosquitoes, Anopheles gambiae and Anopheles coluzzii [J]. Proc Natl Acad Sci USA, 2023, 120(29):e2221118120. DOI:10.1073/pnas.2221118120 . |
| [27] | GANTZ V M, JASINSKIENE N, TATARENKOVA O, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi [J]. Proc Natl Acad Sci USA, 2015, 112(49):E6736-E6743. DOI:10.1073/pnas.1521077112 . |
| [28] | MUÑOZ D, JIMENEZ A, MARINOTTI O, et al. The AeAct-4 gene is expressed in the developing flight muscles of female Aedes aegypti [J]. Insect Mol Biol, 2004, 13(5):563-568. DOI:10.1111/j.0962-1075.2004.00519.x . |
| [29] | O'LEARY S, ADELMAN Z N. CRISPR/Cas9 knockout of female-biased genes AeAct-4 or myo-fem in Ae. aegypti results in a flightless phenotype in female, but not male mosquitoes[J]. PLoS Negl Trop Dis, 2020, 14(12):e0008971. DOI:10.1371/journal.pntd.0008971 . |
| [30] | FRANZ A W E, SANCHEZ-VARGAS I, ADELMAN Z N, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti [J]. Proc Natl Acad Sci USA, 2006, 103(11):4198-4203. DOI:10.1073/pnas.0600479103 . |
| [31] | WILLIAMS A E, SANCHEZ-VARGAS I, REID W R, et al. The antiviral small-interfering RNA pathway induces zika virus resistance in transgenic Aedes aegypti [J]. Viruses, 2020, 12(11):1231. DOI:10.3390/v12111231 . |
| [32] | BUCHMAN A, GAMEZ S, LI M, et al. Broad dengue neutralization in mosquitoes expressing an engineered antibody[J]. PLoS Pathog, 2020, 16(1):e1008103. DOI:10.1371/journal.ppat.1008103 . |
| [33] | REID W R, OLSON K E, FRANZ A W E. Current effector and gene-drive developments to engineer arbovirus-resistant Aedes aegypti (Diptera: Culicidae) for a sustainable population replacement strategy in the field[J]. J Med Entomol, 2021, 58(5):1987-1996. DOI:10.1093/jme/tjab030 . |
| [34] | LI M, YANG T, KANDUL N P, et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti [J]. eLife, 2020, 9:e51701. DOI:10.7554/eLife.51701 . |
| [35] | FENG X C, LÓPEZ DEL AMO V, MAMELI E, et al. Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes[J]. Nat Commun, 2021, 12(1):2960. DOI:10.1038/s41467-021-23239-0 . |
| [36] | FENG X C, KAMBIC L, NISHIMOTO J H K, et al. Evaluation of gene knockouts by CRISPR as potential targets for the genetic engineering of the mosquito Culex quinquefasciatus [J]. CRISPR J, 2021, 4(4):595-608. DOI:10.1089/crispr.2021.0028 . |
| [37] | HARVEY-SAMUEL T, FENG X C, OKAMOTO E M, et al. CRISPR-based gene drives generate super-Mendelian inheritance in the disease vector Culex quinquefasciatus [J]. Nat Commun, 2023, 14(1):7561. DOI:10.1038/s41467-023-41834-1 . |
| [38] | KANDUL N P, LIU J R, BUCHMAN A, et al. Assessment of a split homing based gene drive for efficient knockout of multiple genes[J]. G3 (Bethesda), 2020, 10(2):827-837. DOI:10.1534/g3.119.400985 . |
| [39] | CHAMPER J, REEVES R, OH S Y, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations[J]. PLoS Genet, 2017, 13(7):e1006796. DOI:10.1371/journal.pgen.1006796 . |
| [40] | NOBLE C, ADLAM B, CHURCH G M, et al. Current CRISPR gene drive systems are likely to be highly invasive in wild populations[J]. eLife, 2018, 7:e33423. DOI:10.7554/eLife.33423 . |
| [41] | PROWSE T A A, CASSEY P, ROSS J V, et al. Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates[J]. Proc Biol Sci, 2017, 284(1860):20170799. DOI: 10.1098/rspb.2017.0799 . |
| [42] | CHAMPER J, LIU J X, OH S Y, et al. Reducing resistance allele formation in CRISPR gene drive[J]. Proc Natl Acad Sci USA, 2018, 115(21):5522-5527. DOI:10.1073/pnas.1720354115 . |
| [43] | VELLA M R, GUNNING C E, LLOYD A L, et al. Evaluating strategies for reversing CRISPR-Cas9 gene drives[J]. Sci Rep, 2017, 7(1):11038. DOI: 10.1038/s41598-017-10633-2 . |
| [44] | ESVELT K M, SMIDLER A L, CATTERUCCIA F, et al. Concerning RNA-guided gene drives for the alteration of wild populations[J]. eLife, 2014, 3:e03401. DOI: 10.7554/eLife.03401 . |
| [45] | WU B, LUO L Q, GAO X J. Cas9-triggered chain ablation of cas9 as a gene drive brake[J]. Nat Biotechnol, 2016, 34(2):137-138. DOI:10.1038/nbt.3444 . |
| [46] | ADOLFI A, GANTZ V M, JASINSKIENE N, et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi [J]. Nat Commun, 2020, 11(1):5553. DOI: 10.1038/s41467-020-19426-0 . |
| [47] | SÁNCHEZ C H M, WU S L, BENNETT J B, et al. MGDrivE: a modular simulation framework for the spread of gene drives through spatially explicit mosquito populations[J]. Meth Ecol Evol, 2020, 11(2):229-239. DOI:10.1111/2041-210X.13318 . |
| [48] | NORTH A R, BURT A, GODFRAY H C J. Modelling the suppression of a malaria vector using a CRISPR-Cas9 gene drive to reduce female fertility[J]. BMC Biol, 2020, 18(1):98. DOI: 10.1186/s12915-020-00834-z . |
| [49] | OBERHOFER G, IVY T, HAY B A. Gene drive that results in addiction to a temperature-sensitive version of an essential gene triggers population collapse in Drosophila [J]. Proc Natl Acad Sci USA, 2021, 118(49):e2107413118. DOI: 10.1073/pnas.2107413118 . |
| [50] | WANG G D, VEGA-RODRÍGUEZ J, DIABATE A, et al. Clock genes and environmental cues coordinate Anopheles pheromone synthesis, swarming, and mating[J]. Science, 2021, 371(6527):411-415. DOI:10.1126/science.abd4359 . |
| [1] | 梁敏, 郭洋, 王津津, 朱梦妍, 池骏, 陈艳娟, 王成稷, 喻智澜, 沈如凌. Dmd基因突变小鼠构建及在肌肉及免疫系统的表型验证[J]. 实验动物与比较医学, 2024, 44(1): 42-51. |
| [2] | 李思迪, 付彬, 郭中坤, 林颖杰, 张振宇, 米传靓, 王可洲. 利用CRISPR/Cas9技术构建脂多糖结合蛋白基因敲除小鼠[J]. 实验动物与比较医学, 2022, 42(4): 294-300. |
| [3] | 赖梭梅, 丁一夫, 李劲松. 复杂疾病小鼠模型构建新策略:类精子单倍体胚胎干细胞介导的半克隆技术[J]. 实验动物与比较医学, 2021, 41(5): 369-383. |
| [4] | 洪胜辉, 张旭亮, 王芊芊, 刘 平, 刘迪文. 抑制素基因敲除小鼠模型的构建及表型初步分析[J]. 实验动物与比较医学, 2020, 40(4): 306-. |
| [5] | 高梦樵, 艾东旭, 李钰, 孙菲, 王进, 范君文, 袁征, 刘源, 孙兆增. 利用CRISPR/Cas9技术构建环指蛋白126基因敲除小鼠[J]. 实验动物与比较医学, 2019, 39(1): 21-25. |
| [6] | 张麒, 王建飞. 异种器官移植的进展及展望[J]. 实验动物与比较医学, 2018, 38(6): 407-411. |
| [7] | 艾东旭, 钟德刚, 孙菲, 李钰, 李崇, 秦佟童, 高梦樵, 董施施, 孙兆增, 李莲瑞. 利用CRISPR/Csa9技术构建溶酶体运输调节因子基因缺陷C57BL/6小鼠[J]. 实验动物与比较医学, 2018, 38(3): 202-206. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||