
Laboratory Animal and Comparative Medicine ›› 2024, Vol. 44 ›› Issue (4): 357-373.DOI: 10.12300/j.issn.1674-5817.2024.008
• Animal Models of Human Diseases • Previous Articles Next Articles
WU Yue1( ), LI Lu2, ZHANG Yang2, WANG Jue1, FENG Tingting1, LI Yitong1, WANG Kai2, KONG Qi1(
), LI Lu2, ZHANG Yang2, WANG Jue1, FENG Tingting1, LI Yitong1, WANG Kai2, KONG Qi1( )(
)( )
)
Received:2024-01-16
Revised:2024-04-10
Online:2024-08-25
Published:2024-09-06
Contact:
KONG Qi
CLC Number:
WU Yue,LI Lu,ZHANG Yang,et al. Integrative Analysis of Omics Data in Animal Models of Coronavirus Infection[J]. Laboratory Animal and Comparative Medicine, 2024, 44(4): 357-373. DOI: 10.12300/j.issn.1674-5817.2024.008.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2024.008
数据库名称 Database name | 网址 URL | 简介 Brief description |
|---|---|---|
| Biostudies | https://www.ebi.ac.uk/biostudies/ | 包含生物学研究数据集,以及与EMBL-EBI数据库或其他公共数据库的数据链接 |
| ENA (European Nucleotide Archive) | https://www.ebi.ac.uk/ena | 核酸测序原始数据、序列拼装和功能注释信息 |
| PRIDE (Proteomics Identifications Database) | https://www.ebi.ac.uk/pride | 蛋白质组学质谱数据存储平台包含蛋白质鉴定、翻译后修饰和光谱数据 |
| GEO (Gene Expression Omnibus) | https://www.ncbi.nlm.nih.gov/geo/ | 存储各种高通量实验数据的公共数据库,以表达谱、芯片数据为主 |
| EGA (European Genome-Phenome Archive) | https://ega-archive.org | 存储生物医学研究中产生的遗传和表型数据 |
| ArrayExpress | https://www.ebi.ac.uk/arrayexpress/ | 主要包括微阵列芯片和高通量测序数据,与GEO数据库类似 |
| iProX (Integrated Proteome Resources) | https://www.iprox.org | 国家蛋白质科学中心网站,收录蛋白质组学相关数据集 |
MassIVE (Mass Spectrometry Interactive Virtual Environment) | https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp | 储存蛋白、多肽、质谱数据 |
| FAIRDOMHub | https://fairdomhub.org/ | 共享多种类型的科学研究数据集、模型或研究过程和成果数据 |
| BioModels | https://www.ebi.ac.uk/biomodels/ | 储存生物和医学相关的数学模型、生理学和药学相关机制模型 |
| MetaboLights | https://www.ebi.ac.uk/metabolights/ | 储存代谢组学和相关衍生信息的数据库,包含代谢物的结构、浓度、功能数据 |
| dbGap (Database of Genotypes and Phenotypes) | https://dbgap.ncbi.nlm.nih.gov/ | 储存基因型和表型互作数据 |
| PanoramaPublic | https://panoramaweb.org/ | 储存蛋白质组学和小分子数据 |
| GNPS (Global Natural Products Social Molecular Networking) | https://gnps.ucsd.edu/ | 储存原始、处理或注释过的质谱数据 |
Metabolomics Workbench | https://www.metabolomicsworkbench.org/ | 代谢组学元数据和实验数据的公共数据库,包含不同物种的质谱(MS)和核磁共振(NMR)光谱数据 |
| ExpressionAtlas | https://www.ebi.ac.uk/gxa/home | 提供不同条件下基因表达信息,包含芯片与转录组数据 |
jPOST (Japan Proteome Standard Repository Database) | https://jpostdb.org/ | 包含蛋白质组数据及分析结果数据 |
| EVA (European Variation Archive) | https://www.ebi.ac.uk/eva/ | 包含不同物种遗传变异数据 |
| NODE (National Omics Data Encyclopedia) | https://www.biosino.org/node/ | 储存多组学数据资源,包括测序数据、蛋白质组学数据、代谢组学数据及荧光成像数据 |
| PeptideAtlas | https://peptideatlas.org/ | 收录质谱、蛋白质组学数据。主要为人、小鼠、酵母数据 |
Table 1 Public databases containing coronavirus omics data
数据库名称 Database name | 网址 URL | 简介 Brief description |
|---|---|---|
| Biostudies | https://www.ebi.ac.uk/biostudies/ | 包含生物学研究数据集,以及与EMBL-EBI数据库或其他公共数据库的数据链接 |
| ENA (European Nucleotide Archive) | https://www.ebi.ac.uk/ena | 核酸测序原始数据、序列拼装和功能注释信息 |
| PRIDE (Proteomics Identifications Database) | https://www.ebi.ac.uk/pride | 蛋白质组学质谱数据存储平台包含蛋白质鉴定、翻译后修饰和光谱数据 |
| GEO (Gene Expression Omnibus) | https://www.ncbi.nlm.nih.gov/geo/ | 存储各种高通量实验数据的公共数据库,以表达谱、芯片数据为主 |
| EGA (European Genome-Phenome Archive) | https://ega-archive.org | 存储生物医学研究中产生的遗传和表型数据 |
| ArrayExpress | https://www.ebi.ac.uk/arrayexpress/ | 主要包括微阵列芯片和高通量测序数据,与GEO数据库类似 |
| iProX (Integrated Proteome Resources) | https://www.iprox.org | 国家蛋白质科学中心网站,收录蛋白质组学相关数据集 |
MassIVE (Mass Spectrometry Interactive Virtual Environment) | https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp | 储存蛋白、多肽、质谱数据 |
| FAIRDOMHub | https://fairdomhub.org/ | 共享多种类型的科学研究数据集、模型或研究过程和成果数据 |
| BioModels | https://www.ebi.ac.uk/biomodels/ | 储存生物和医学相关的数学模型、生理学和药学相关机制模型 |
| MetaboLights | https://www.ebi.ac.uk/metabolights/ | 储存代谢组学和相关衍生信息的数据库,包含代谢物的结构、浓度、功能数据 |
| dbGap (Database of Genotypes and Phenotypes) | https://dbgap.ncbi.nlm.nih.gov/ | 储存基因型和表型互作数据 |
| PanoramaPublic | https://panoramaweb.org/ | 储存蛋白质组学和小分子数据 |
| GNPS (Global Natural Products Social Molecular Networking) | https://gnps.ucsd.edu/ | 储存原始、处理或注释过的质谱数据 |
Metabolomics Workbench | https://www.metabolomicsworkbench.org/ | 代谢组学元数据和实验数据的公共数据库,包含不同物种的质谱(MS)和核磁共振(NMR)光谱数据 |
| ExpressionAtlas | https://www.ebi.ac.uk/gxa/home | 提供不同条件下基因表达信息,包含芯片与转录组数据 |
jPOST (Japan Proteome Standard Repository Database) | https://jpostdb.org/ | 包含蛋白质组数据及分析结果数据 |
| EVA (European Variation Archive) | https://www.ebi.ac.uk/eva/ | 包含不同物种遗传变异数据 |
| NODE (National Omics Data Encyclopedia) | https://www.biosino.org/node/ | 储存多组学数据资源,包括测序数据、蛋白质组学数据、代谢组学数据及荧光成像数据 |
| PeptideAtlas | https://peptideatlas.org/ | 收录质谱、蛋白质组学数据。主要为人、小鼠、酵母数据 |
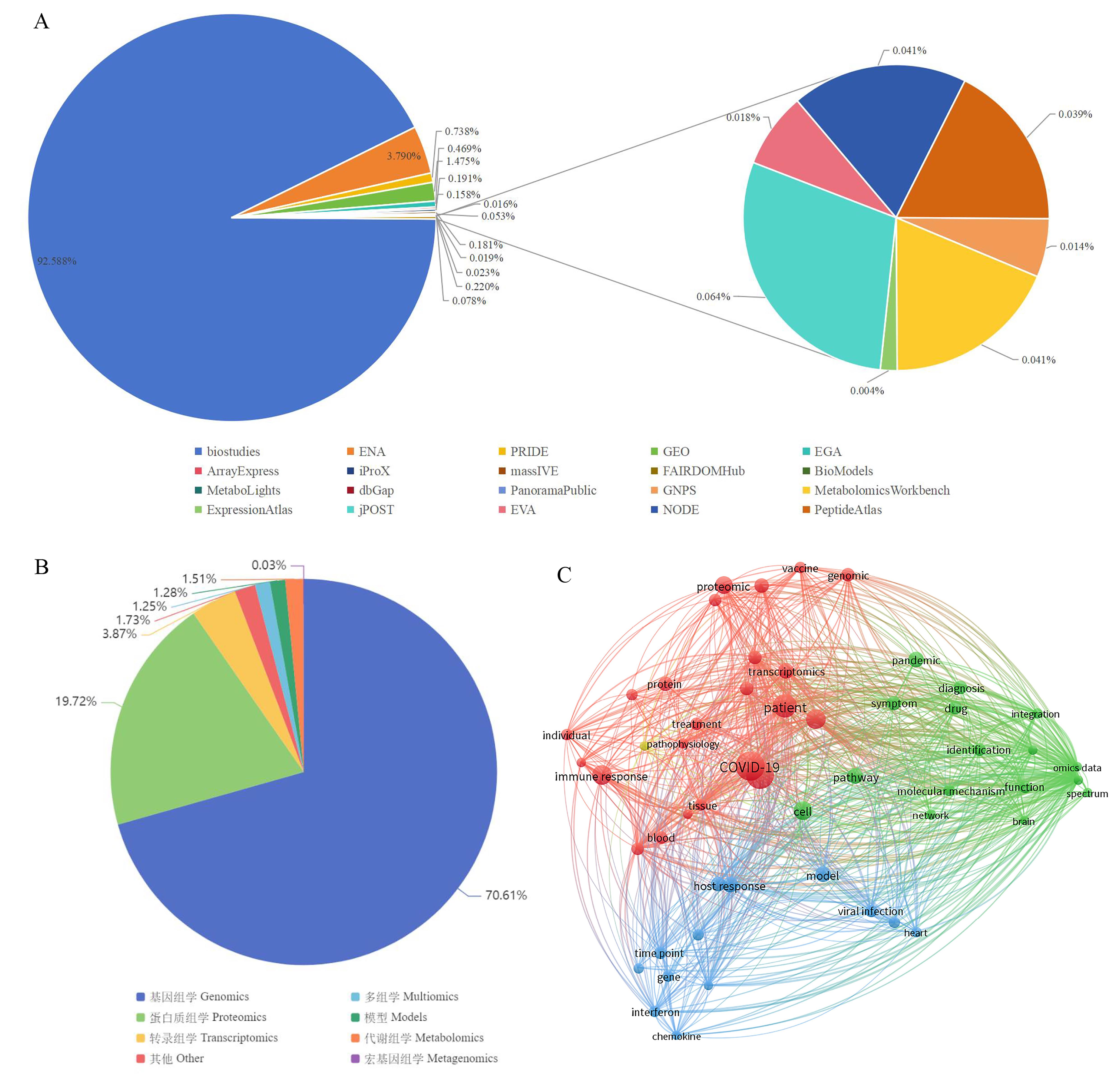
Figure 1 Statistics and text co-occurrence network analysis of coronavirus multi-omics data in public databasesNote:A, Distribution of coronavirus omics data in public databases; B, Data types of SARS-CoV-2 in OmicsDI; C, Text co-occurrence network analysis of multi-omics related to SARS-CoV-2.
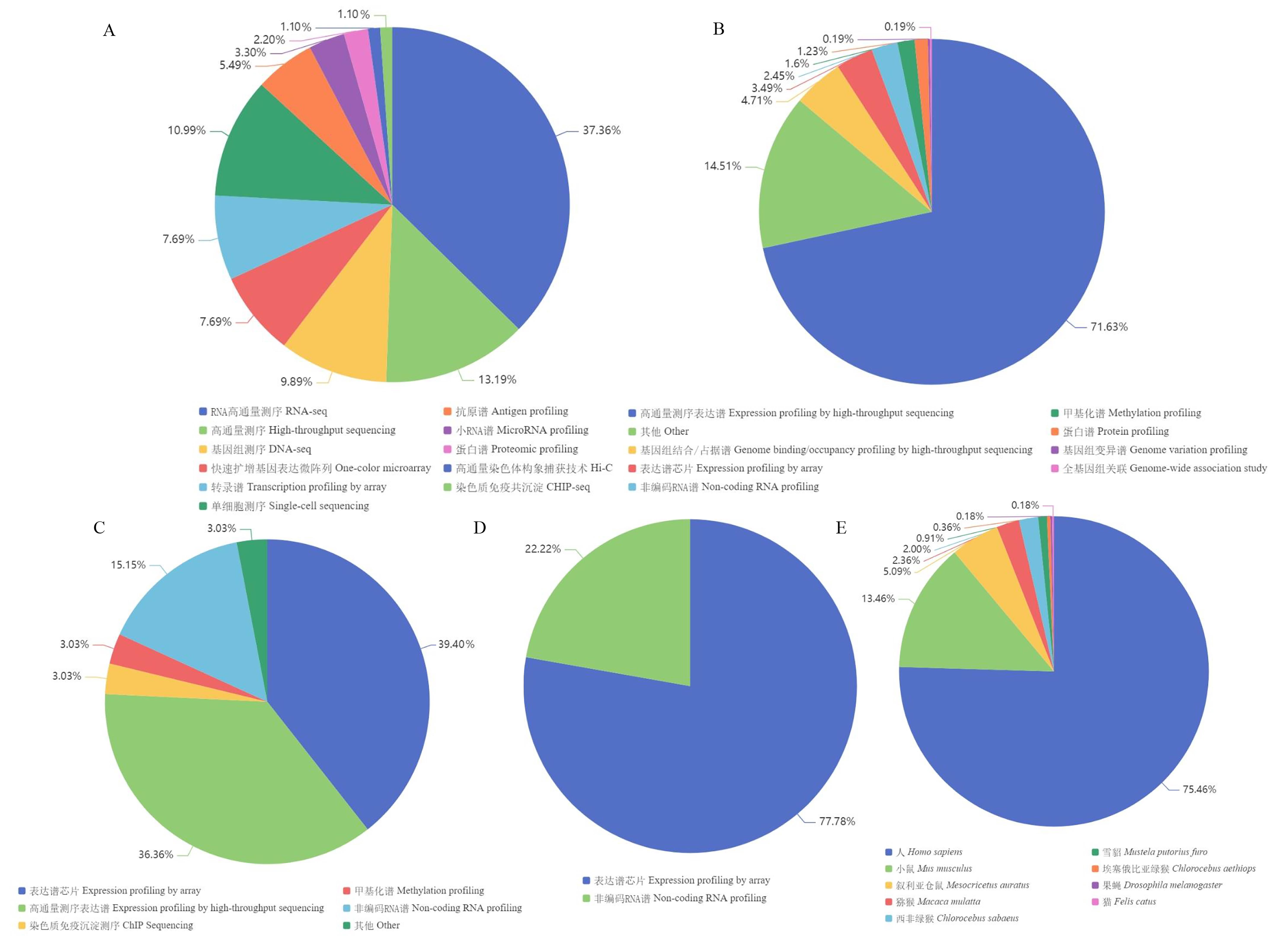
Figure 2 Statistics and analysis of coronavirus omics data in ArrayExpress, GEO, and DDBJNote:A, Data types of SARS-CoV-2 in ArrayExpress; B, Data types of SARS-CoV-2 in GEO; C, Data types of MERS in GEO; D, Data types of SARS in GEO; E, Species distribution of SARS-CoV-2 data in DDBJ.
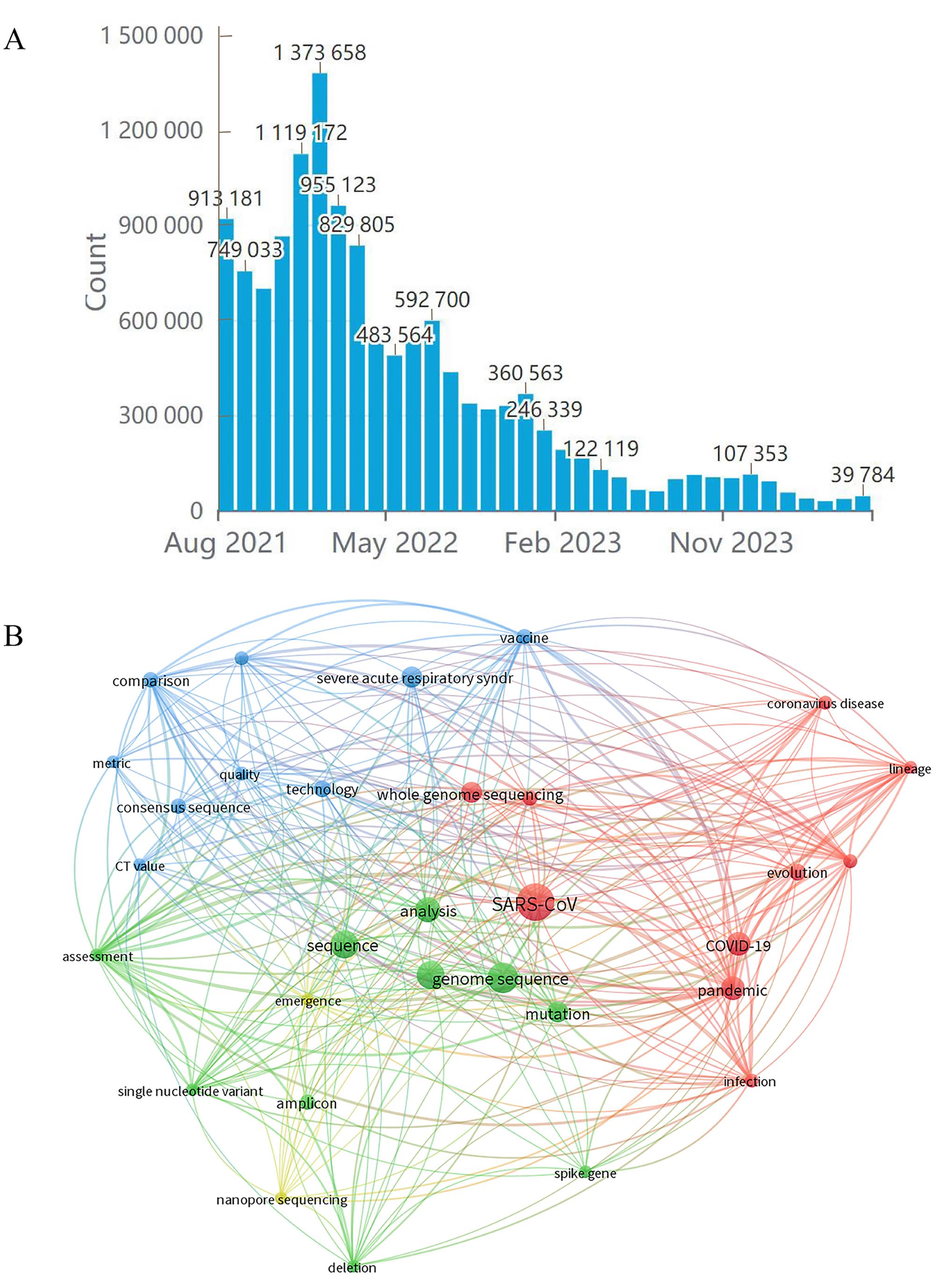
Figure 3 Statistics and analysis of SARS-CoV-2 genomics dataNote: A, Changes of SARS-CoV-2 sequence data in GISAID; B, Text co-occurrence network analysis of SARS-CoV-2 genomics.
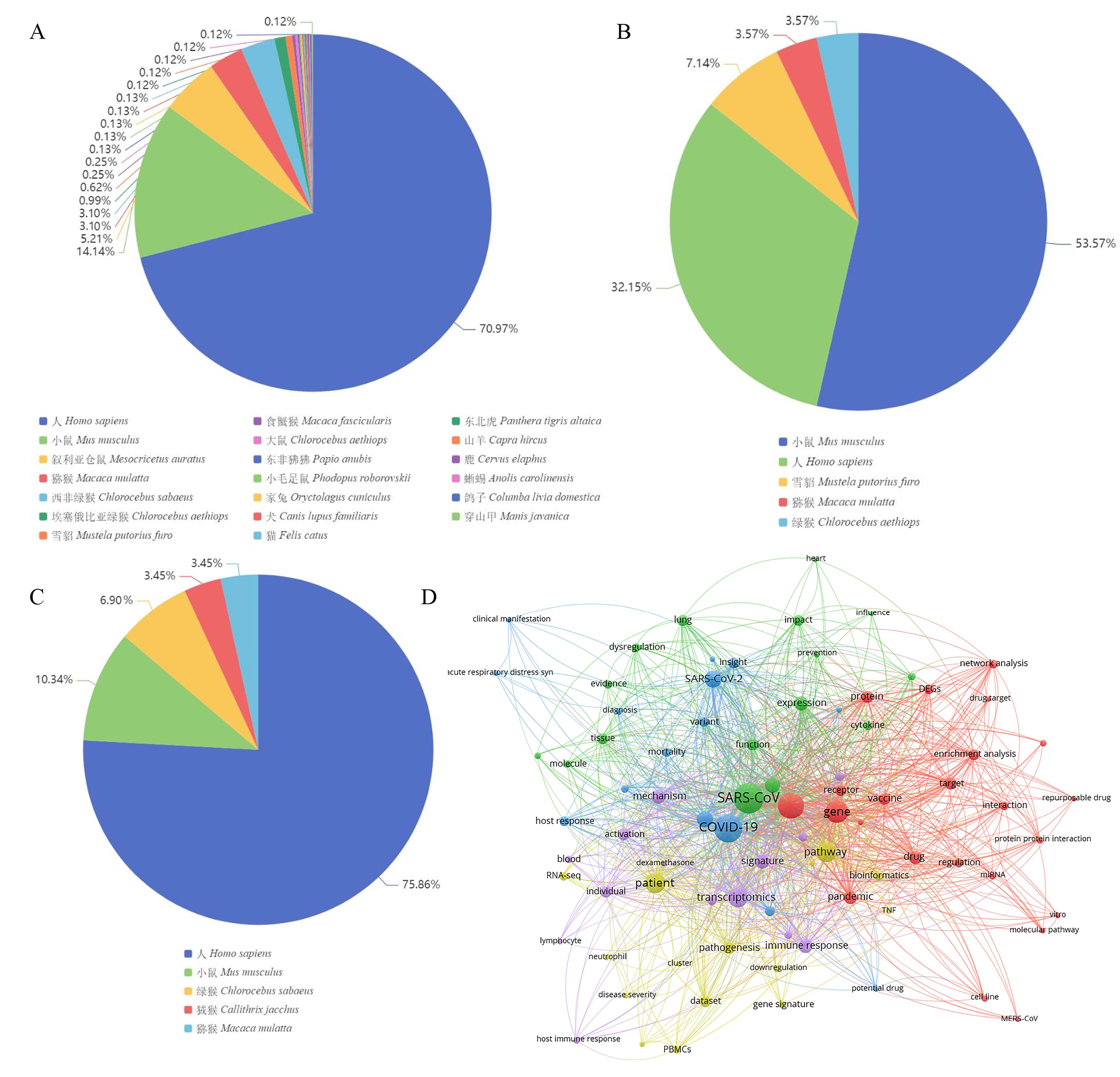
Figure 4 Statistics and analysis of coronavirus expression profiles in GEONote: A, Species distribution of SARS-CoV-2 expression profiles in GEO; B, Species distribution of SARS expression profiles in GEO; C, Species distribution of MERS expression profiles in GEO; D, Text co-occurrence network analysis of SARS-CoV-2 in transcriptomics.
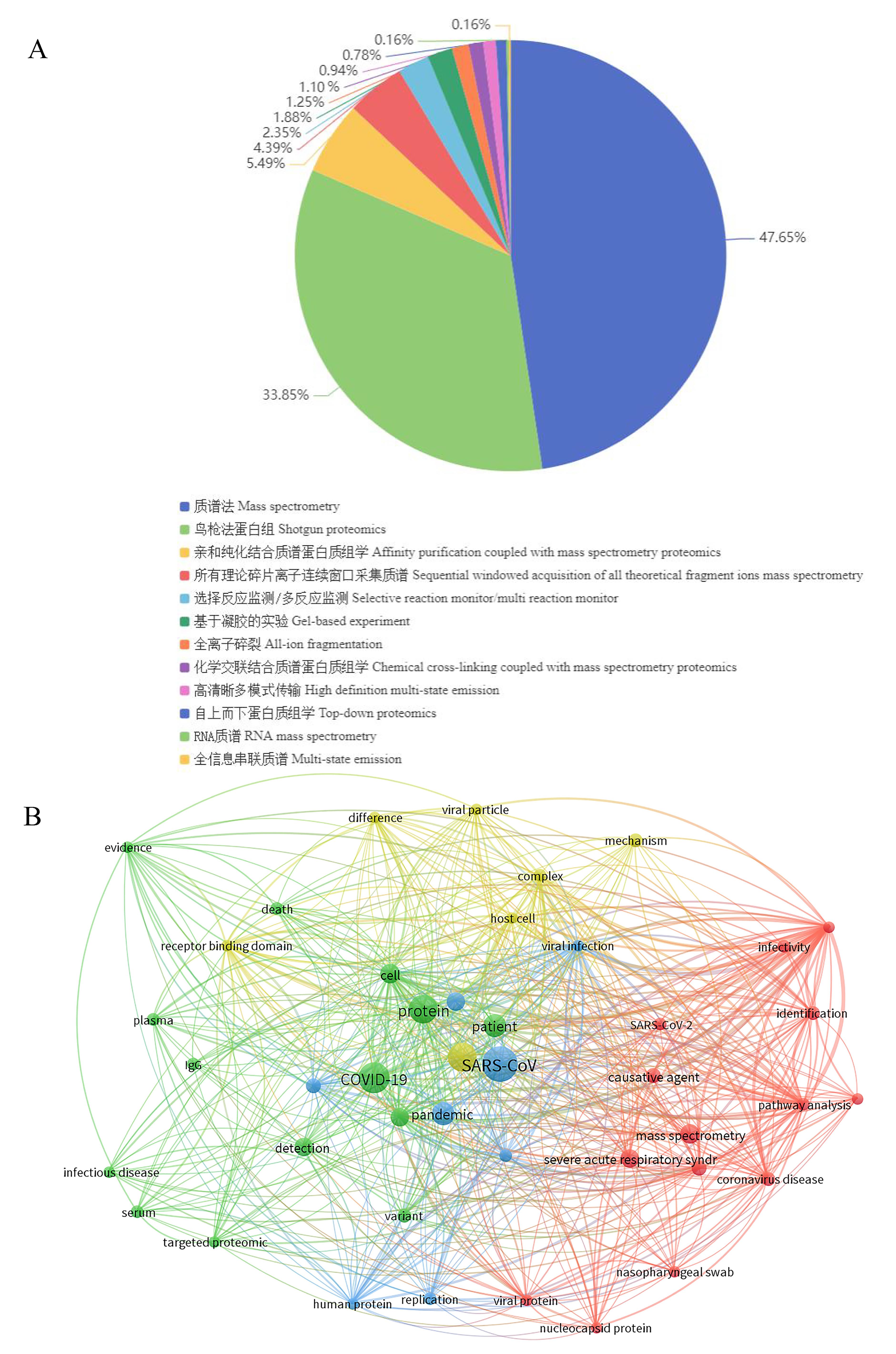
Figure 5 Statistics and text co-occurrence networkNote: A,Types of coronavirus proteomics data; B, Text co-occurrence network analysis of SARS-CoV-2 proteomics.analysis of coronavirus proteomics data
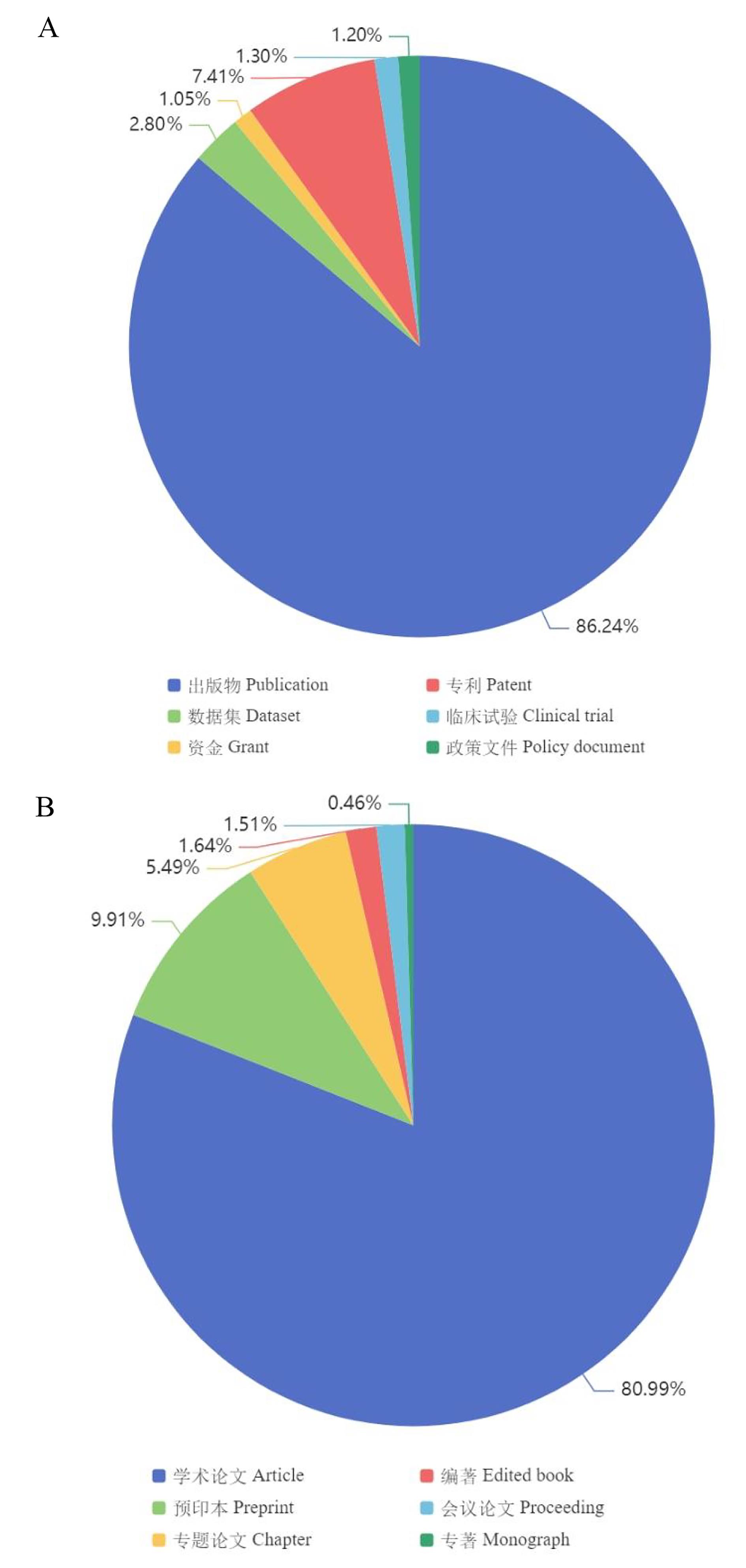
Figure 7 Distribution of coronavirus-related data and literatureNote:A, Types of coronavirus-related data in Dimension; B, Types of coronavirus-related publications in Dimension.
| 1 | KHALIFA S A M, MOHAMED B S, ELASHAL M H, et al. Comprehensive overview on multiple strategies fighting COVID-19[J]. Int J Environ Res Public Health, 2020, 17(16):5813. DOI: 10.3390/ijerph17165813 . |
| 2 | SHEHATA M M, GOMAA M R, ALI M A, et al. Middle East respiratory syndrome coronavirus: a comprehensive review[J]. Front Med, 2016, 10(2):120-136. DOI: 10.1007/s11684-016-0430-6 . |
| 3 | RANA R, TRIPATHI A, KUMAR N, et al. A comprehensive overview on COVID-19: future perspectives[J]. Front Cell Infect Microbiol, 2021, 11:744903. DOI: 10.3389/fcimb. 2021.744903 . |
| 4 | CHAN J F, KOK K H, ZHU Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan[J]. Emerg Microbes Infect, 2020, 9(1):221-236. DOI: 10.1080/22221751.2020.1719902 . |
| 5 | SHI W F, LI J, ZHOU H, et al. Pathogen genomic surveillance elucidates the origins, transmission and evolution of emerging viral agents in China[J]. Sci China Life Sci, 2017, 60(12):1317-1330. DOI: 10.1007/s11427-017-9211-0 . |
| 6 | DAS J K, TRADIGO G, VELTRI P, et al. Data science in unveiling COVID-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing[J]. Brief Bioinform, 2021, 22(2):855-872. DOI: 10.1093/bib/bbaa420 . |
| 7 | SAYERS E W, BECK J, BOLTON E E, et al. Database resources of the National Center for Biotechnology Information[J]. Nucleic Acids Res, 2024, 52(D1): D33-D43. DOI: 10.1093/nar/gkad1044 . |
| 8 | O'SULLIVAN C, BUSBY B, MIZRACHI I K. Managing sequence data[J]. Methods Mol Biol, 2017, 1525:79-106. DOI: 10.1007/978-1-4939-6622-6_4 . |
| 9 | SHUMWAY M, COCHRANE G, SUGAWARA H. Archiving next generation sequencing data[J]. Nucleic Acids Res, 2010, 38(Database issue): D870-D871. DOI: 10.1093/nar/gkp1078 . |
| 10 | WILKINSON M D, DUMONTIER M, AALBERSBERG I JAN, et al. Addendum: the FAIR Guiding Principles for scientific data management and stewardship[J]. Sci Data, 2019, 6(1):6. DOI: 10.1038/s41597-019-0009-6 . |
| 11 | MONTALDO C, MESSINA F, ABBATE I, et al. Multi-omics approach to COVID-19: a domain-based literature review[J]. J Transl Med, 2021, 19(1):501. DOI: 10.1186/s12967-021-03168-8 . |
| 12 | VENKATAKRISHNAN A J, PURANIK A, ANAND A, et al. Knowledge synthesis of 100 million biomedical documents augments the deep expression profiling of coronavirus receptors[J]. Elife, 2020, 9: e58040. DOI: 10.7554/eLife.58040 . |
| 13 | KOLENC Ž, PIRIH N, GRETIC P, et al. Top trends in multiomics research: evaluation of 52 published studies and new ways of thinking terminology and visual displays[J]. OMICS, 2021, 25(11):681-692. DOI: 10.1089/omi.2021.0160 . |
| 14 | 周大琼, 曹继华, 任力锋, 等. 基因芯片数据库GEO与ArrayExpress的使用及比较分析[J]. 中国现代医学杂志, 2014, 24(12):38-42. DOI:10.3969/j.issn.1005-8982.2014.12.009 . |
| ZHOU D Q, CAO J H, REN L F, et al. Usage and comparative analysis of GEO and ArrayExpress microarray database[J]. China J Mod Med, 2014, 24(12):38-42. DOI:10.3969/j.issn.1005- 8982.2014.12.009 . | |
| 15 | MURILLO J, VILLEGAS L M, ULLOA-MURILLO L M, et al. Recent trends on omics and bioinformatics approaches to study SARS-CoV-2: a bibliometric analysis and mini-review[J]. Comput Biol Med, 2021, 128:104162. DOI: 10.1016/j.compbiomed.2020.104162 . |
| 16 | VAN ECK N J, WALTMAN L. Software survey: VOSviewer, a computer program for bibliometric mapping[J]. Scientometrics, 2010, 84(2):523-538. DOI: 10.1007/s11192-009-0146-3 . |
| 17 | DASS G, VU M T, XU P, et al. The omics discovery REST interface[J]. Nucleic Acids Res, 2020, 48(W1): W380-W384. DOI: 10.1093/nar/gkaa326 . |
| 18 | BARH D, TIWARI S, WEENER M E, et al. Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19[J]. Comput Biol Med, 2020, 126:104051. DOI: 10.1016/j.compbiomed.2020.104051 . |
| 19 | GUO M Y, XIONG M Y, PENG J Y, et al. Multi-omics for COVID-19: driving development of therapeutics and vaccines[J]. Natl Sci Rev, 2023, 10(9): nwad161. DOI: 10.1093/nsr/nwad161 . |
| 20 | DRAKE K A, TALANTOV D, TONG G J, et al. Multi-omic profiling reveals early immunological indicators for identifying COVID-19 Progressors[J]. Clin Immunol, 2023, 256:109808. DOI: 10.1016/j.clim.2023.109808 . |
| 21 | LI C X, GAO J, ZHANG Z C, et al. Multiomics integration-based molecular characterizations of COVID-19[J]. Brief Bioinform, 2022, 23(1): bbab485. DOI: 10.1093/bib/bbab485 . |
| 22 | SARAVANAN K A, PANIGRAHI M, KUMAR H, et al. Role of genomics in combating COVID-19 pandemic[J]. Gene, 2022, 823:146387. DOI: 10.1016/j.gene.2022.146387 . |
| 23 | ROUCHKA E C, CHARIKER J H, CHUNG D. Variant analysis of 1,040 SARS-CoV-2 genomes[J]. PLoS One, 2020, 15(11): e0241535. DOI: 10.1371/journal.pone.0241535 . |
| 24 | GAO C C, LI M, DENG W, et al. Differential transcriptomic landscapes of multiple organs from SARS-CoV-2 early infected rhesus macaques[J]. Protein Cell, 2022, 13(12):920-939. DOI: 10.1007/s13238-022-00915-5 . |
| 25 | RUAN Q R, YANG K, WANG W X, et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China[J]. Intensive Care Med, 2020, 46(5):846-848. DOI: 10.1007/s00134-020-05991-x . |
| 26 | KIM J A, KIM S H, SEO J S, et al. Temporal transcriptome analysis of SARS-CoV-2-infected lung and spleen in human ACE2-transgenic mice[J]. Mol Cells, 2022, 45(12):896-910. DOI: 10.14348/molcells.2022.0089 . |
| 27 | DALY J L, SIMONETTI B, KLEIN K, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection[J]. Science, 2020, 370(6518):861-865. DOI: 10.1126/science.abd3072 . |
| 28 | O'DONNELL K L, PINSKI A N, CLANCY C S, et al. Pathogenic and transcriptomic differences of emerging SARS-CoV-2 variants in the Syrian golden hamster model[J]. EBioMedicine, 2021, 73:103675. DOI: 10.1016/j.ebiom. 2021. 103675 . |
| 29 | DESAI N, NEYAZ A, SZABOLCS A, et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection[J]. Nat Commun, 2020, 11(1):6319. DOI: 10.1038/s41467-020-20139-7 . |
| 30 | OH T, KIM G, BAEK S H, et al. Comparative spatial transcriptomic profiling of severe acute respiratory syndrome coronavirus 2 Delta and Omicron variants infections in the lungs of cynomolgus macaques[J]. J Med Virol, 2023, 95(6): e28847. DOI: 10.1002/jmv.28847 . |
| 31 | ESPOSITO S, D'ABROSCA G, ANTOLAK A, et al. Host and viral zinc-finger proteins in COVID-19[J]. Int J Mol Sci, 2022, 23(7):3711. DOI: 10.3390/ijms23073711 . |
| 32 | GUPTA R K, ROSENHEIM J, BELL L C, et al. Blood transcriptional biomarkers of acute viral infection for detection of pre-symptomatic SARS-CoV-2 infection: a nested, case-control diagnostic accuracy study[J]. Lancet Microbe, 2021, 2(10): e508-e517. DOI: 10.1016/S2666-5247(21)00146-4 . |
| 33 | ZIEGLER C G K, MIAO V N, OWINGS A H, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19[J]. Cell, 2021, 184(18):4713-4733.e22. DOI: 10.1016/j.cell.2021.07.023 . |
| 34 | NIE X, QIAN L J, SUN R, et al. Multi-organ proteomic landscape of COVID-19 autopsies[J]. Cell, 2021, 184(3):775-791.e14. DOI: 10.1016/j.cell.2021.01.004 . |
| 35 | MESSNER C B, DEMICHEV V, WENDISCH D, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection[J]. Cell Syst, 2020, 11(1):11-24.e4. DOI: 10.1016/j.cels.2020.05.012 . |
| 36 | BOJKOVA D, KLANN K, KOCH B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets[J]. Nature, 2020, 583(7816):469-472. DOI: 10.1038/s41586-020-2332-7 . |
| 37 | XU J J, ZHOU M, LUO P, et al. Plasma metabolomic profiling of patients recovered from coronavirus disease 2019 (COVID-19) with pulmonary sequelae 3 months after discharge[J]. Clin Infect Dis, 2021, 73(12):2228-2239. DOI: 10.1093/cid/ciab147 . |
| 38 | SHEN B, YI X, SUN Y T, et al. Proteomic and metabolomic characterization of COVID-19 patient sera[J]. Cell, 2020, 182(1):59-72.e15. DOI: 10.1016/j.cell.2020.05.032 . |
| 39 | KARPE A V, NGUYEN T V, SHAH R M, et al. A time-series metabolomic analysis of SARS-CoV-2 infection in a ferret model[J]. Metabolites, 2022, 12(11):1151. DOI: 10.3390/metabo12111151 . |
| 40 | JIA H L, LIU C W, LI D T, et al. Metabolomic analyses reveal new stage-specific features of COVID-19[J]. Eur Respir J, 2022, 59(2):2100284. DOI: 10.1183/13993003.00284-2021 . |
| 41 | VALDÉS A, MORENO L O, RELLO S R, et al. Metabolomics study of COVID-19 patients in four different clinical stages[J]. Sci Rep, 2022, 12(1):1650. DOI: 10.1038/s41598-022-05667-0 . |
| 42 | THOMAS T, STEFANONI D, REISZ J A, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status[J]. JCI Insight, 2020, 5(14): e140327. DOI: 10.1172/jci.insight.140327 . |
| 43 | MA J L, DENG Y W, ZHANG M H, et al. The role of multi-omics in the diagnosis of COVID-19 and the prediction of new therapeutic targets[J]. Virulence, 2022, 13(1):1101-1110. DOI: 10.1080/21505594.2022.2092941 . |
| 44 | KGATLE M M, LAWAL I O, MASHABELA G, et al. COVID-19 is a multi-organ aggressor: epigenetic and clinical marks[J]. Front Immunol, 2021, 12:752380. DOI: 10.3389/fimmu. 2021.752380 . |
| 45 | BALNIS J, MADRID A, HOGAN K J, et al. Persistent blood DNA methylation changes one year after SARS-CoV-2 infection[J]. Clin Epigenetics, 2022, 14(1):94. DOI: 10.1186/s13148-022-01313-8 . |
| 46 | STUKALOV A, GIRAULT V, GRASS V, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV[J]. Nature, 2021, 594(7862):246-252. DOI: 10.1038/s41586-021-03493-4 . |
| 47 | PHILLIPS S, MISHRA T, KHADKA S, et al. Epitranscriptomic N 6-methyladenosine profile of SARS-CoV-2-infected human lung epithelial cells[J]. Microbiol Spectr, 2023, 11(1): e0394322. DOI: 10.1128/spectrum.03943-22 . |
| 48 | MA S L, ZHANG F, ZHOU F X, et al. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19[J]. Signal Transduct Target Ther, 2021, 6(1):191. DOI: 10.1038/s41392-021-00614-3 . |
| 49 | MAEDA Y, MOTOOKA D, KAWASAKI T, et al. Longitudinal alterations of the gut mycobiota and microbiota on COVID-19 severity[J]. BMC Infect Dis, 2022, 22(1):572. DOI: 10.1186/s12879-022-07358-7 . |
| 50 | ZUO T, ZHANG F, LUI G C Y, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization[J]. Gastroenterology, 2020, 159(3):944-955.e8. DOI: 10.1053/j.gastro.2020.05.048 . |
| 51 | LIU Q, MAK J W Y, SU Q, et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome[J]. Gut, 2022, 71(3):544-552. DOI: 10.1136/gutjnl-2021-325989 . |
| 52 | NIES L D, GALATA V, MARTIN-GALLAUSIAUX C, et al. Altered infective competence of the human gut microbiome in COVID-19[J]. Microbiome, 2023, 11(1):46. DOI: 10.1186/s40168-023-01472-7 . |
| 53 | XU X G, ZHANG W, GUO M Q, et al. Integrated analysis of gut microbiome and host immune responses in COVID-19[J]. Front Med, 2022, 16(2):263-275. DOI: 10.1007/s11684-022-0921-6 . |
| 54 | PANG Z Q, ZHOU G Y, CHONG J, et al. Comprehensive meta-analysis of COVID-19 global metabolomics datasets[J]. Metabolites, 2021, 11(1):44. DOI: 10.3390/metabo11010044 . |
| 55 | LU T, WANG Y R, GUO T N. Multi-omics in COVID-19: seeing the unseen but overlooked in the clinic[J]. Cell Rep Med, 2022, 3(3):100580. DOI: 10.1016/j.xcrm.2022.100580 . |
| 56 | CEN X P, WANG F G, HUANG X H, et al. Towards precision medicine: Omics approach for COVID-19[J]. Biosaf Health, 2023, 5(2):78-88. DOI: 10.1016/j.bsheal.2023.01.002 . |
| 57 | MIRALLES A, BRUY T, WOLCOTT K, et al. Repositories for taxonomic data: where we are and what is missing[J]. Syst Biol, 2020, 69(6):1231-1253. DOI: 10.1093/sysbio/syaa026 . |
| 58 | HAAS P, MURALIDHARAN M, KROGAN N J, et al. Proteomic approaches to study SARS-CoV-2 biology and COVID-19 pathology[J]. J Proteome Res, 2021, 20(2):1133-1152. DOI: 10.1021/acs.jproteome.0c00764 . |
| 59 | YANG J J, YAN Y Z, ZHONG W. Application of omics technology to combat the COVID-19 pandemic[J]. MedComm, 2021, 2(3):381-401. DOI: 10.1002/mco2.90 . |
| 60 | RAY S, SRIVASTAVA S. COVID-19 pandemic: hopes from proteomics and multiomics research[J]. OMICS, 2020, 24(8):457-459. DOI: 10.1089/omi.2020.0073 . |
| 61 | ZHU Z J, ZHANG S N, WANG P, et al. A comprehensive review of the analysis and integration of omics data for SARS-CoV-2 and COVID-19[J]. Brief Bioinform, 2022, 23(1): bbab446. DOI: 10.1093/bib/bbab446 . |
| 62 | DONG X J, LIU C Y, DOZMOROV M. Review of multi-omics data resources and integrative analysis for human brain disorders[J]. Brief Funct Genomics, 2021, 20(4):223-234. DOI: 10.1093/bfgp/elab024 . |
| 63 | 吴玥, 向志光, 高苒, 等. 冠状病毒感染动物模型比较转录组学数据库的建立[J]. 中国实验动物学报, 2022, 30(1):92-99. DOI: 10.3969/j.issn.1005-4847.2022.01.012 . |
| WU Y, XIANG Z G, GAO R, et al. Establishment of a comparative transcriptomics database of coronavirus infected animal models[J]. Acta Lab Anim Sci Sin, 2022, 30(1):92-99. DOI: 10.3969/j.issn.1005-4847.2022.01.012 . | |
| 64 | 吴玥, 王珏, 薛婧, 等. 心脏病动物模型比较转录组学数据库的构建[J]. 中国比较医学杂志, 2023, 33(3):75-81. DOI: 10.3969/j.issn.1671-7856.2023.03.010 . |
| WU Y, WANG J, XUE J, et al. Establishment of a comparative transcriptomics database of heart disease animal models[J]. Chin J Comp Med, 2023, 33(3):75-81. DOI: 10.3969/j.issn.1671-7856.2023.03.010 . | |
| 65 | 孔琪, 夏霞宇, 秦川. 实验动物品系数据库的建立[J]. 中国比较医学杂志, 2015, 25(4):78-83. DOI: 10.3969/j.issn.1671.7856.2015.004.016 . |
| KONG Q, XIA X Y, QIN C. Establishment of the laboratory animal strain database[J]. Chin J Comp Med, 2015, 25(4):78-83. DOI: 10.3969/j.issn.1671.7856.2015.004.016 . | |
| 66 | 吴玥, 魏强, 张连峰, 等. 比较医学大数据平台的建立[J]. 中国比较医学杂志, 2022, 32(12):57-65. DOI: 10.3969/j.issn.1671-7856.2022.12.008 . |
| WU Y, WEI Q, ZHANG L F, et al. Establishment the comparative medicine big-data platform[J]. Chin J Comp Med, 2022, 32(12):57-65. DOI: 10.3969/j.issn.1671-7856.2022.12.008 . | |
| 67 | 吴玥, 薛婧, 魏强, 等. 国家动物模型资源共享信息平台的建立[J]. 中国实验动物学报, 2022, 30(8):1080-1086. DOI: 10.3969/j.issn.1005-4847.2022.08.009 . |
| WU Y, XUE J, WEI Q, et al. Establishment of national infrastructure for an animal model resource platform[J]. Acta Lab Anim Sci Sin, 2022, 30(8):1080-1086. DOI: 10.3969/j.issn.1005-4847.2022.08.009 . | |
| 68 | 吴玥, 王珏, 冯婷婷, 等. 基于动物模型的药物筛选数据库的建立[J]. 中国实验动物学报, 2023, 31(6):778-786. DOI: 10.3969/j.issn.1005-4847.2023.06.010 . |
| WU Y, WANG J, FENG T T, et al. Construction of a drug screening database based on animal models[J]. Acta Lab Anim Sci Sin, 2023, 31(6):778-786. DOI: 10.3969/j.issn.1005-4847.2023.06.010 . | |
| 69 | WU Y, WANG J, XUE J, et al. Flu-CED: A comparative transcriptomics database of influenza virus-infected human and animal models[J]. Animal Model Exp Med,2024,2:2-12. DOI: 10.1002/ame2.12384 . |
| [1] | LIU Yayi, JIA Yunfeng, ZUO Yiming, ZHANG Junping, LÜ Shichao. Progress and Evaluation of Animal Model of Heart Qi-Yin Deficiency Syndrome [J]. Laboratory Animal and Comparative Medicine, 2025, 45(4): 411-421. |
| [2] | PAN Yicong, JIANG Wenhong, HU Ming, QIN Xiao. Optimization of Surgical Procedure and Efficacy Evaluation of Aortic Calcification Model in Rats with Chronic Kidney Disease [J]. Laboratory Animal and Comparative Medicine, 2025, 45(3): 279-289. |
| [3] | LIAN Hui, JIANG Yanling, LIU Jia, ZHANG Yuli, XIE Wei, XUE Xiaoou, LI Jian. Construction and Evaluation of a Rat Model of Abnormal Uterine Bleeding [J]. Laboratory Animal and Comparative Medicine, 2025, 45(2): 130-146. |
| [4] | YANG Jiahao, DING Chunlei, QIAN Fenghua, SUN Qi, JIANG Xusheng, CHEN Wen, SHEN Mengwen. Research Progress on Animal Models of Sepsis-Related Organ Injury [J]. Laboratory Animal and Comparative Medicine, 2024, 44(6): 636-644. |
| [5] | HUANG Dongyan, WU Jianhui. Establishment Methods and Application Evaluation of Animal Models in Reproductive Toxicology Research [J]. Laboratory Animal and Comparative Medicine, 2024, 44(5): 550-559. |
| [6] | ZHENG Yiqing, DENG Yasheng, FAN Yanping, LIANG Tianwei, HUANG Hui, LIU Yonghui, NI Zhaobing, LIN Jiang. Application Analysis of Animal Models for Pelvic Inflammatory Disease Based on Data Mining [J]. Laboratory Animal and Comparative Medicine, 2024, 44(4): 405-418. |
| [7] | Committee of Experts on Medical Animal Experiments, Chinese Research Hospital Association. Guidelines for the Selection of Animal Models and Preclinical Drug Trials for Spontaneous Intracerebral Hemorrhage (2024 Edition) [J]. Laboratory Animal and Comparative Medicine, 2024, 44(1): 3-30. |
| [8] | Shuwu XIE, Ruling SHEN, Jinxing LIN, Chun FAN. Progress in Establishment and Application of Laboratory Animal Models Related to Development of Male Infertility Drugs [J]. Laboratory Animal and Comparative Medicine, 2023, 43(5): 504-511. |
| [9] | Rui ZHANG, Meiyu LÜ, Jianjun ZHANG, Jinlian LIU, Yan CHEN, Zhiqiang HUANG, Yao LIU, Lanhua ZHOU. Research Progress on Establishing and Evaluation of Acne Animal Models [J]. Laboratory Animal and Comparative Medicine, 2023, 43(4): 398-405. |
| [10] | Jiahui YU, Qian GONG, Lenan ZHUANG. Animal Models of Pulmonary Arterial Hypertension and Their Application in Drug Research [J]. Laboratory Animal and Comparative Medicine, 2023, 43(4): 381-397. |
| [11] | Ling HU, Zhibin HU, Yunqing HU, Yuqiang DING. Overview of Studies in Animal Models of Schizophrenia [J]. Laboratory Animal and Comparative Medicine, 2023, 43(2): 145-155. |
| [12] | Feng WEI, Weiwei CHENG, Yafu YIN. Characteristics and Application of Transgenic Mouse Models in Alzheimer's Disease [J]. Laboratory Animal and Comparative Medicine, 2022, 42(5): 432-439. |
| [13] | Zhejin SHENG, Limei LI. Research Progress in Animal Model of Alzheimer's Disease [J]. Laboratory Animal and Comparative Medicine, 2022, 42(4): 342-350. |
| [14] | Yaohua HU, Jumei ZHAO, Changhong SHI. Application of Animal Models in the Functional Studies of Eph Family Proteins [J]. Laboratory Animal and Comparative Medicine, 2022, 42(4): 351-357. |
| [15] | Xiao LI, Haipeng YAN, Zhenghui XIAO. Construction Methods and Influencing Factors on Animal Model of Sepsis [J]. Laboratory Animal and Comparative Medicine, 2022, 42(3): 207-212. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||