













• •
出版日期:2025-10-28
作者简介:岳君佳俞(1998-),博士研究生,研究方向:整合生命科学。E-mail: yjjy@stu.pku.edu.cn。ORCID: 0000-0001-8685-5664;基金资助:Published:2025-10-28
Contact:
ZHANG Wei (ORCID: 0000-0002-6644-7046), E-mail: weizhangvv@pku.edu.cn摘要:
理解动物多样性的起源及其适应性演化机制,是现代生物学的核心问题之一。动物形态、体色、行为等性状由物种的遗传和发育以及环境因素协同塑造,其相互作用机制长期受到广泛关注。高通量测序和基因编辑等技术的发展,促进了非模式生物的演化生物学研究。蝶类(Lepidoptera: Papilionoidea)因其分布广泛、翅图案多样化、生活史短以及便于人工饲养等特点,已成为研究动物演化的重要模式生物。近年来,为了揭示多种复杂性状的适应性演化规律,关于蝶类拟态及其遗传机制、季节型与表型可塑性、以及环境感知与互作等方面的研究不断深入。本文总结了当前在蝶类遗传、发育与演化层面的主要研究体系和方向,包括多种蝶类类群适应性演化的经典体系,例如:(1)doublesex (dsx) 及相关基因决定了凤蝶属(Papilio)雌性特异的贝氏拟态。(2)optix基因调控元件的改变,驱动了袖蝶属(Heliconius)不同物种间翅图案的趋同演化,为揭示穆氏拟态的演化提供了分子层面的直接证据。(3)以枯叶蛱蝶属(Kallima)为例,其叶形拟态由包含cortex基因的基因组区域所控制。除拟态性状外,本文也概述了蝶类旱雨季型的表型可塑性,鹿眼眼蝶(Junonia coenia)与美眼蛱蝶(Bicyclus anynana)是研究环境适应与发育可塑性的经典模型。此外,蝶类还是研究复杂生物—环境互作的重要体系,包括柑橘凤蝶(Papilio xuthus)的视觉感知与色彩识别机制、菜粉蝶(Pieris rapae)与寄主植物协同演化,以及君主斑蝶(Danaus plexippus)的长距离迁飞与警戒色相关机制等。本文揭示了蝶类演化研究中整合多组学数据(如泛基因组、单细胞转录组等)解析基因调控网络的研究趋势。本文还从技术发展和研究方向拓展的角度对蝶类研究进行了展望,为动物演化研究提供了新思路。综上所述,蝶类研究体系形成了整合遗传、发育、环境互作及行为模拟的多学科交叉新范式。这一范式不仅深化了对生物适应性演化规律的理解,并提供了可延伸的研究框架,推动了演化生物学与生态保护、工程应用以及生理疾病等领域的融合与创新。
中图分类号:
岳君佳俞,张蔚.蝶类演化研究体系的构建与分析进展[J]. 实验动物与比较医学.. DOI: 10.12300/j.issn.1674-5817.2025.099.
YUE Junjiayu,ZHANG Wei. Progress in Constructing and Analyzing the Evolutionary Research Framework for Butterflies[J]. Laboratory Animal and Comparative Medicine. DOI: 10.12300/j.issn.1674-5817.2025.099.
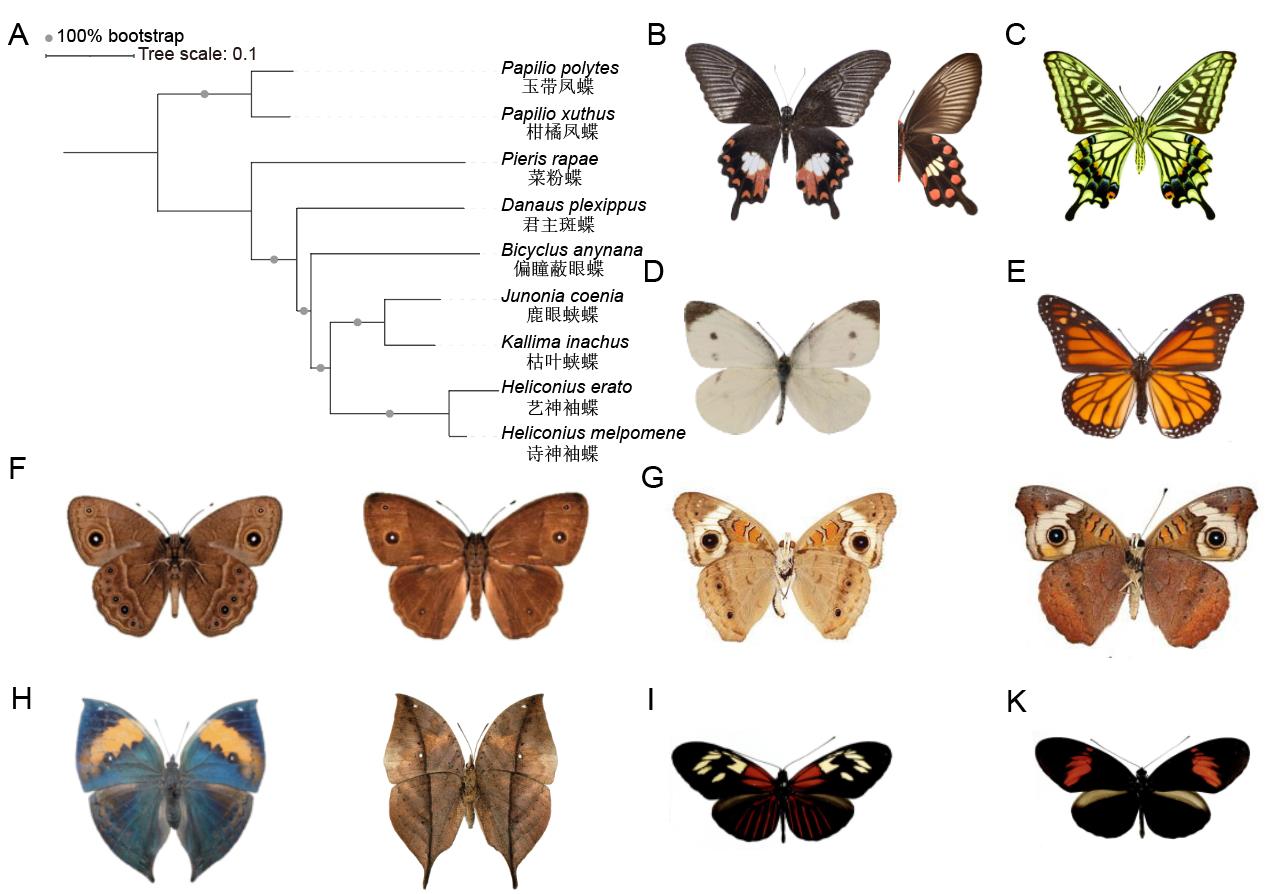
图1 代表性蝶类研究类群的系统发生关系及其翅表型注:A,基于OrthoFinder鉴定了9个物种的直系同源基因,使用2,711个单拷贝基因构建了最大似然树;B,玉带凤蝶雌性贝氏拟态个体翅背侧,右为其拟态对象红珠凤蝶翅背侧;C,柑橘凤蝶翅背侧;D,菜粉蝶翅背侧;E,君主斑蝶翅背侧;F,偏瞳蔽眼蝶翅背侧,左为雨季型,右为旱季型;G,鹿眼蛱蝶翅背侧,左为雨季型,右为旱季型;H,枯叶蛱蝶,左为翅背侧,右为其叶形伪装拟态的翅腹侧;I,艺神袖蝶翅背侧;K,诗神袖蝶翅背侧。
Figure 1 The phylogeny and the wing pattern of the representative butterfly research taxaNote:A, Orthologous genes from nine species were identified using OrthoFinder, and a maximum likelihood phylogenetic tree was constructed based on 2,711 single-copy genes; B, the dorsal side of a mimetic female Papilio polytes, with the dorsal side of its mimetic model Pachliopta aristolochiae on the right; C, the dorsal side of Papilio xuthus; D, the dorsal side of Pieris rapae; E, the dorsal side of Danaus plexippus; F, the dorsal sides of Bicyclus anynana, with the wet seasonal morph on the left and the dry seasonal morph on the right; G, the dorsal sides of Junonia coenia, with the wet seasonal morph on the left and the dry seasonal morph on the right; H, the dorsal side of Kallima inachus is on the left and its ventral side showing leaf wing pattern is on the right; I, the dorsal side of Heliconius erato; K, the dorsal side of Heliconius melpomene.
基因/基因座 Gene/gene locus | 基因功能 Gene function | 蝶类物种 Species | 相关表型 Phenotypes |
|---|---|---|---|
doublesex | 转录因子 | 玉带凤蝶 (Papilio polytes) | 贝氏拟态 [ |
美凤蝶 (Papilio memnon) | 贝氏拟态 [ | ||
红斑美凤蝶 (Papilio rumanzovia) | 贝氏拟态 [ | ||
果园美凤蝶 (Papilio aegeus) | 贝氏拟态 [ | ||
optix | 转录因子 | 袖蝶属(Heliconius) | 穆氏拟态 [ |
银纹红袖蝶 (Agraulis vanillae) | 橙色和红色翅图案 [ | ||
鹿眼蛱蝶 (Junonia coenia) | |||
小红蛱蝶 (Vanessa cardui) | |||
cortex | 细胞周期调控因子 | 枯叶蛱蝶属 (Kallima) | 叶形拟态多样性 [ |
鹿眼蛱蝶 (Junonia coenia) | 季节性表型可塑性 [ | ||
斑凤蝶 (Papilio clytia) | 贝氏拟态 [ | ||
狐眼袖蝶 (Heliconius numata) | 穆氏拟态 [ | ||
| Dll | 转录因子 | 偏瞳蔽眼蝶 (Bicyclus anynana) | 眼斑 [ |
表1 蝶翅花纹研究中的热点基因示例
Table 1 Hotspot genes in butterfly wing pattern research
基因/基因座 Gene/gene locus | 基因功能 Gene function | 蝶类物种 Species | 相关表型 Phenotypes |
|---|---|---|---|
doublesex | 转录因子 | 玉带凤蝶 (Papilio polytes) | 贝氏拟态 [ |
美凤蝶 (Papilio memnon) | 贝氏拟态 [ | ||
红斑美凤蝶 (Papilio rumanzovia) | 贝氏拟态 [ | ||
果园美凤蝶 (Papilio aegeus) | 贝氏拟态 [ | ||
optix | 转录因子 | 袖蝶属(Heliconius) | 穆氏拟态 [ |
银纹红袖蝶 (Agraulis vanillae) | 橙色和红色翅图案 [ | ||
鹿眼蛱蝶 (Junonia coenia) | |||
小红蛱蝶 (Vanessa cardui) | |||
cortex | 细胞周期调控因子 | 枯叶蛱蝶属 (Kallima) | 叶形拟态多样性 [ |
鹿眼蛱蝶 (Junonia coenia) | 季节性表型可塑性 [ | ||
斑凤蝶 (Papilio clytia) | 贝氏拟态 [ | ||
狐眼袖蝶 (Heliconius numata) | 穆氏拟态 [ | ||
| Dll | 转录因子 | 偏瞳蔽眼蝶 (Bicyclus anynana) | 眼斑 [ |
| [1] | DARWIN C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life[M]. A reprint of the first edition. London: Watts, 1950. |
| [2] | PROVINE W B. The Origins of Theoretical Population Genetics[M]. Chicago: University of Chicago Press, 1971. |
| [3] | FISHER R A. XV.: the correlation between relatives on the supposition of mendelian inheritance[J]. Trans R Soc Edinb, 1919, 52(2):399-433. DOI:10.1017/s0080456800012163 . |
| [4] | HALL B K. Evolutionary developmental biology (evo-devo): past, present, and future[J]. Evol Educ Outreach, 2012, 5(2):184-193. DOI:10.1007/s12052-012-0418-x . |
| [5] | MEYER A. Hox gene variation and evolution[J]. Nature, 1998, 391(6664):227-228. DOI:10.1038/34530 . |
| [6] | MONTEIRO A, MURUGESAN S N, PRAKASH A, et al. The developmental origin of novel complex morphological traits in Lepidoptera[J]. Annu Rev Entomol, 2025, 70(1):421-439. DOI:10.1146/annurev-ento-021324-020504 . |
| [7] | CARROLL S B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution[J]. Cell, 2008, 134(1):25-36. DOI:10.1016/j.cell.2008.06.030 . |
| [8] | WAGNER G P, LYNCH V J. Evolutionary novelties[J]. Curr Biol, 2010, 20(2): R48-R52. DOI:10.1016/j.cub.2009.11.010 . |
| [9] | CECH T R, STEITZ J A. The noncoding RNA revolution-trashing old rules to forge new ones[J]. Cell, 2014, 157(1):77-94. DOI:10.1016/j.cell.2014.03.008 . |
| [10] | KUNTE K, ZHANG W, TENGER-TROLANDER A, et al. Doublesex is a mimicry supergene[J]. Nature, 2014, 507(7491):229-232. DOI:10.1038/nature13112 . |
| [11] | MALLET J, JORON M. Evolution of diversity in warning color and mimicry: polymorphisms, shifting balance, and speciation[J]. Annu Rev Ecol Syst, 1999, 30 (1999):201-233. |
| [12] | WANG S T, TENG D Q, LI X Y, et al. The evolution and diversification of oakleaf butterflies[J]. Cell, 2022, 185(17):3138-3152. DOI:10.1016/j.cell.2022.06.042 . |
| [13] | BRAKEFIELD P M, LARSEN T B. The evolutionary significance of dry and wet season forms in some tropical butterflies[J]. Biol J Linn Soc, 1984, 22(1):1-12. DOI:10.1111/j.1095-8312.1984.tb00795.x . |
| [14] | SMITH K C. The effects of temperature and daylength on the Rosa polyphenism in the buckeye butterfly, Precis coenia (Lepidoptera: Nymphalidae)[J]. J Res Lepid, 1993, 30(3-4):225-236. DOI:10.5962/p.266647 . |
| [15] | KINOSHITA M, ARIKAWA K. Colour constancy in the swallowtail butterfly Papilio Xuthus[J]. J Exp Biol, 2000, 203(23):3521-3530. DOI:10.1242/jeb.203.23.3521 . |
| [16] | WHEAT C W, VOGEL H, WITTSTOCK U, et al. The genetic basis of a plant-insect coevolutionary key innovation[J]. Proc Natl Acad Sci USA, 2007, 104(51):20427-20431. DOI:10.1073/pnas.0706229104 . |
| [17] | URQUHART F A, URQUHART N R. Autumnal migration routes of the eastern population of the monarch butterfly (Danaus p. plexippus L.; Danaidae; Lepidoptera) in North America to the overwintering site in the Neovolcanic Plateau of Mexico[J]. Can J Zool, 1978, 56(8):1759-1764. DOI:10.1139/z78-240 . |
| [18] | REICHSTEIN T, VON EUW J, PARSONS J A, et al. Heart Poisons in the Monarch Butterfly: Some aposematic butterflies obtain protection from cardenolides present in their food plants[J]. Science, 1968, 161(3844):861-866. DOI:10.1126/science.161.3844.861 . |
| [19] | BELDADE P, BRAKEFIELD P M. The genetics and evo-devo of butterfly wing patterns[J]. Nat Rev Genet, 2002, 3(6):442-452. DOI:10.1038/nrg818 . |
| [20] | SCHWANWITSCH B N. 21. on the ground-plan of wing-pattern in nymphalids and certain other families of the rhopaloeerous Lepidoptera[J]. Proc Zool Soc Lond, 1924, 94(2):509-528. DOI:10.1111/j.1096-3642.1924.tb01511.x . |
| [21] | NIJHOUT H F. The Development and Evolution of Butterfly Wing Patterns[M]. Smithson. Inst., 1991. |
| [22] | REED R D, SELEGUE J E, ZHANG L L, et al. Transcription factors underlying wing margin color patterns and pupal cuticle markings in butterflies[J]. Evodevo, 2020, 11:10. DOI:10.1186/s13227-020-00155-w . |
| [23] | TIONG G J L, NAING L, NG E, et al. Tympanal ears mediate male-male competition, courtship and mating success in Bicyclus anynana butterflies[J]. R Soc Open Sci, 2024, 11(3):231386. DOI:10.1098/rsos.231386 . |
| [24] | POULTON E B. The colors of animals: their meaning and use, especially considered in the case of insects[M]. London: Kegan Paul, Trench, Trübner & Co., 1890 |
| [25] | BATES H W. XXXII. contributions to an insect fauna of the Amazon valley. Lepidoptera: Heliconidae[J]. Trans Linn Soc Lond, 1862, 23(3):495-566. DOI:10.1111/j.1096-3642.1860.tb00146.x . |
| [26] | CLARKE C A, SHEPPARD P M. Super-genes and mimicry[J]. Heredity, 1960, 14(1-2):175-185. DOI:10.1038/hdy.1960.15 . |
| [27] | JORON M, MALLET J L. Diversity in mimicry: paradox or paradigm?[J]. Trends Ecol Evol, 1998, 13(11):461-466. DOI:10.1016/s0169-5347(98)01483-9 . |
| [28] | CLARKE C A, SHEPPARD P M, THORNTON I W B. The genetics of the mimetic butterfly Papilio memnon L[J]. Philos Trans R Soc Lond Ser B Biol Sci, 1968, 254(791):37-89. |
| [29] | OHSAKI N. Preferential predation of female butterflies and the evolution of batesian mimicry[J]. Nature, 1995, 378(6553):173-175. DOI:10.1038/378173a0 . |
| [30] | KUNTE K. The diversity and evolution of Batesian mimicry in Papilio swallowtail butterflies[J]. Evolution, 2009, 63(10):2707-2716. DOI:10.1111/j.1558-5646.2009.00752.x . |
| [31] | YODA S, SAKAKURA K, KITAMURA T, et al. Genetic switch in UV response of mimicry-related pale-yellow colors in Batesian mimic butterfly, Papilio polytes[J]. Sci Adv, 2021, 7(2): eabd6475. DOI:10.1126/sciadv.abd6475 . |
| [32] | IIJIMA T, KAJITANI R, KOMATA S, et al. Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon[J]. Sci Adv, 2018, 4(4): eaao5416. DOI:10.1126/sciadv.aao5416 . |
| [33] | KOMATA S, YODA S, KONDO Y, et al. Functional unit of supergene in female-limited Batesian mimicry of Papilio polytes[J]. Genetics, 2023, 223(2): iyac177. DOI:10.1093/genetics/iyac177 . |
| [34] | VANKUREN N W, SHEIKH S I, FU C L, et al. Functional genetic elements of a butterfly mimicry supergene[J]. Proc Natl Acad Sci USA, 2025, 122(41): e2509864122. DOI:10.1073/pnas.2509864122 . |
| [35] | PALMER D H, KRONFORST M R. A shared genetic basis of mimicry across swallowtail butterflies points to ancestral co-option of doublesex[J]. Nat Commun, 2020, 11(1):6. DOI:10.1038/s41467-019-13859-y . |
| [36] | KOMATA S, KAJITANI R, ITOH T, et al. Genomic architecture and functional unit of mimicry supergene in female limited Batesian mimic Papilio butterflies[J]. Philos Trans R Soc Lond B Biol Sci, 2022, 377(1856):20210198. DOI:10.1098/rstb.2021.0198 . |
| [37] | MüLLER F. Ituna and Thyridia: a remarkable case of mimicry in butterflies[J]. Trans Entomol Soc Lond, 1879: 20-29. |
| [38] | BENSON W W. Resource partitioning in passion vine butterflies[J]. Evolution, 1978, 32(3):493-518. DOI:10.1111/j.1558-5646.1978.tb04593.x . |
| [39] | MALLET J, GILBERT L E Jr. Why are there so many mimicry rings? Correlations between habitat, behaviour and mimicry in Heliconius butterflies[J]. Biol J Linn Soc, 1995, 55(2):159-180. DOI:10.1111/j.1095-8312.1995.tb01057.x . |
| [40] | TURNER J R G. Studies of müllerian mimicry and its evolution in burnet moths and heliconid butterflies[M]//Ecological Genetics and Evolution. New York, NY: Springer US, 1971:224-260. DOI:10.1007/978-1-4757-0432-7_11 . |
| [41] | PAPA R, MARTIN A, REED R D. Genomic hotspots of adaptation in butterfly wing pattern evolution[J]. Curr Opin Genet Dev, 2008, 18(6):559-564. DOI:10.1016/j.gde.2008.11.007 . |
| [42] | REED R D, PAPA R, MARTIN A, et al. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry[J]. Science, 2011, 333(6046):1137-1141. DOI:10.1126/science.1208227 . |
| [43] | WALLACE A R. Darwinism: AN EXPOSITION OF THE THEORY OF NATURAL SELECTION, WITH SOME OF ITS APPLICATIONS[M]. New York: Macmillan, 1889. |
| [44] | TENG D Q, LI F Y, ZHANG W. Using comprehensive machine-learning models to classify complex morphological characters[J]. Ecol Evol, 2021, 11(15):10421-10431. DOI:10.1002/ece3.7845 . |
| [45] | ZHANG Z T, YU L, CHANG H Z, et al. Nature's disguise: Empirical demonstration of dead-leaf masquerade in Kallima butterflies[J]. Zool Res, 2024, 45(6):1201-1208. DOI:10.24272/j.issn.2095-8137.2024.025 . |
| [46] | YANG J, WAN W T, XIE M, et al. Chromosome-level reference genome assembly and gene editing of the dead-leaf butterfly Kallima inachus[J]. Mol Ecol Resour, 2020, 20(4):1080-1092. DOI:10.1111/1755-0998.13185 . |
| [47] | VAN DER BURG K R, REED R D. Seasonal plasticity: how do butterfly wing pattern traits evolve environmental responsiveness?[J]. Curr Opin Genet Dev, 2021, 69:82-87. DOI:10.1016/j.gde.2021.02.009 . |
| [48] | BRAKEFIELD P M, GATES J, KEYS D, et al. Development, plasticity and evolution of butterfly eyespot patterns[J]. Nature, 1996, 384(6606):236-242. DOI:10.1038/384236a0 . |
| [49] | OLIVER J C, RAMOS D, PRUDIC K L, et al. Temporal gene expression variation associated with eyespot size plasticity in Bicyclus anynana[J]. PLoS One, 2013, 8(6): 1-5. DOI:10.1371/journal.pone.0065830 . |
| [50] | TIAN S, MONTEIRO A. A transcriptomic atlas underlying developmental plasticity of seasonal forms of Bicyclus anynana butterflies[J]. Mol Biol Evol, 2022, 39(6): msac126. DOI:10.1093/molbev/msac126 . |
| [51] | ROUNTREE D B, NIJHOUT H F. Hormonal control of a seasonal polyphenism in Precis coenia (Lepidoptera: Nymphalidae)[J]. J Insect Physiol, 1995, 41(11):987-992. DOI:10.1016/0022-1910(95)00046-W . |
| [52] | VAN DER BURG K R L, LEWIS J J, BRACK B J, et al. Genomic architecture of a genetically assimilated seasonal color pattern[J]. Science, 2020, 370(6517):721-725. DOI:10.1126/science.aaz3017 . |
| [53] | FANDINO R A, BRADY N K, CHATTERJEE M, et al. The ivory lncRNA regulates seasonal color patterns in buckeye butterflies[J]. Proc Natl Acad Sci USA, 2024, 121(41): 1-8. DOI:10.1073/pnas.2403426121 . |
| [54] | SRYGLEY R B, THOMAS A L R. Unconventional lift-generating mechanisms in free-flying butterflies[J]. Nature, 2002, 420(6916):660-664. DOI:10.1038/nature01223 . |
| [55] | NYLIN S, BERGSTRÖM A, JANZ N. Butterfly host plant choice in the face of possible confusion[J]. J Insect Behav, 2000, 13(4):469-482. DOI:10.1023/A: 1007839200323 . |
| [56] | BROWER L P. Understanding and misunderstanding the migration of the monarch butterfly (Nymphalidae) in North America: 1857-1995[J]. J Lepidopterists Soc, 1995, 49:304-385. |
| [57] | ARIKAWA K. Spectral organization of the eye of a butterfly, Papilio[J]. J Comp Physiol A, 2003, 189(11):791-800. DOI:10.1007/s00359-003-0454-7 . |
| [58] | KINOSHITA M, SHIMADA N, ARIKAWA K. Colour vision of the foraging swallowtail butterfly Papilio xuthus[J]. J Exp Biol, 1999, 202 (Pt 2):95-102. DOI:10.1242/jeb.202.2.95 . |
| [59] | KINOSHITA M, TAKAHASHI Y, ARIKAWA K. Simultaneous color contrast in the foraging swallowtail butterfly, Papilio xuthus[J]. J Exp Biol, 2008, 211(Pt 21):3504-3511. DOI:10.1242/jeb.017848 . |
| [60] | YAMAGUCHI S, WOLF R, DESPLAN C, et al. Motion vision is independent of color in Drosophila[J]. Proc Natl Acad Sci USA, 2008, 105(12):4910-4915. DOI:10.1073/pnas.0711484105 . |
| [61] | STEWART F J, KINOSHITA M, ARIKAWA K. Monopolatic motion vision in the butterfly Papilio xuthus[J]. J Exp Biol, 2019, 222(Pt 1): jeb191957. DOI:10.1242/jeb.191957 . |
| [62] | CÉCHETTO C, ARIKAWA K, KINOSHITA M. Motion-sensitive neurons activated by chromatic contrast in a butterfly visual system[J]. Philos Trans R Soc Lond B Biol Sci, 2022, 377(1862):20210277. DOI:10.1098/rstb.2021.0277 . |
| [63] | MARTIN A, MCCULLOCH K J, PATEL N H, et al. Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations[J]. Evodevo, 2014, 5(1):7. DOI:10.1186/2041-9139-5-7 . |
| [64] | ZHANG L L, MAZO-VARGAS A, REED R D. Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence[J]. Proc Natl Acad Sci USA, 2017, 114(40):10707-10712. DOI:10.1073/pnas.1709058114 . |
| [65] | VANKUREN N W, MASSARDO D, NALLU S, et al. Butterfly mimicry polymorphisms highlight phylogenetic limits of gene reuse in the evolution of diverse adaptations[J]. Mol Biol Evol, 2019, 36(12):2842-2853. DOI:10.1093/molbev/msz194 . |
| [66] | NADEAU N J, PARDO-DIAZ C, WHIBLEY A, et al. A major gene controls mimicry and crypsis in butterflies and moths[J]. Nature, 2016, 534(7605):106-110. DOI:10.1038/nature17961 . |
| [67] | JORON M, FREZAL L, JONES R T, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry[J]. Nature, 2011, 477(7363):203-206. DOI:10.1038/nature10341 . |
| [68] | FAHEY J W, ZALCMANN A T, TALALAY P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants[J]. Phytochemistry, 2001, 56(1):5-51. DOI:10.1016/s0031-9422(00)00316-2 . |
| [69] | WITTSTOCK U, AGERBIRK N, STAUBER E J, et al. Successful herbivore attack due to metabolic diversion of a plant chemical defense[J]. Proc Natl Acad Sci USA, 2004, 101(14):4859-4864. DOI:10.1073/pnas.0308007101 . |
| [70] | SMID H M, VAN LOON J J A, POSTHUMUS M A, et al. GC-EAG-analysis of volatiles from Brussels sprouts plants damaged by two species of Pieris caterpillars: olfactory receptive range of a specialist and a generalist parasitoid wasp species[J]. CHEMOECOLOGY, 2002, 12(4):169-176. DOI:10.1007/PL00012665 . |
| [71] | PUENTE M, MAGORI K, KENNEDY G G, et al. Impact of herbivore-induced plant volatiles on parasitoid foraging success: a spatial simulation of the Cotesia rubecula, Pieris rapae, and Brassica oleracea system[J]. J Chem Ecol, 2008, 34(7):959-970. DOI:10.1007/s10886-008-9472-9 . |
| [72] | BRUINSMA M, POSTHUMUS M A, MUMM R, et al. Jasmonic acid-induced volatiles of Brassica oleracea attract parasitoids: effects of time and dose, and comparison with induction by herbivores[J]. J Exp Bot, 2009, 60(9):2575-2587. DOI:10.1093/jxb/erp101 . |
| [73] | ZHAN S, MERLIN C, BOORE J L, et al. The monarch butterfly genome yields insights into long-distance migration[J]. Cell, 2011, 147(5):1171-1185. DOI:10.1016/j.cell.2011.09.052 . |
| [74] | ZHAN S, ZHANG W, NIITEPÕLD K, et al. The genetics of monarch butterfly migration and warning colouration[J]. Nature, 2014, 514(7522):317-321. DOI:10.1038/nature13812 . |
| [75] | VAN'T HOF A E, CAMPAGNE P, RIGDEN D J, et al. The industrial melanism mutation in British peppered moths is a transposable element[J]. Nature, 2016, 534(7605):102-105. DOI:10.1038/nature17951 . |
| [76] | TANG F C, BARBACIORU C, WANG Y Z, et al. mRNA-Seq whole-transcriptome analysis of a single cell[J]. Nat Methods, 2009, 6(5):377-382. DOI:10.1038/nmeth.1315 . |
| [77] | PRAKASH A, DION E, BANERJEE T DAS, et al. The molecular basis of scale development highlighted by a single-cell atlas of Bicyclus anynana butterfly pupal forewings[J]. Cell Rep, 2024, 43(5):114147. DOI:10.1016/j.celrep.2024.114147 . |
| [78] | LOH L S, DEMARR K A, TSIMBA M, et al. Lepidopteran scale cells derive from sensory organ precursors through a canonical lineage[J]. Development, 2025, 152(5): DEV204501. DOI:10.1242/dev.204501 . |
| [79] | BUENROSTRO J D, GIRESI P G, ZABA L C, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position[J]. Nat Methods, 2013, 10(12):1213-1218. DOI:10.1038/nmeth.2688 . |
| [80] | TIAN S, ASANO Y, BANERJEE T DAS, et al. A microRNA is the effector gene of a classic evolutionary hotspot locus[J]. Science, 2024, 386(6726):1135-1141. DOI:10.1126/science.adp7899 . |
| [81] | LIVRAGHI L, HANLY J J, EVANS E, et al. A long noncoding RNA at the cortex locus controls adaptive coloration in butterflies[J]. Proc Natl Acad Sci USA, 2024, 121(36): e2403326121. DOI:10.1073/pnas.2403326121 . |
| [82] | KARAGEORGI M, GROEN S C, SUMBUL F, et al. Genome editing retraces the evolution of toxin resistance in the monarch butterfly[J]. Nature, 2019, 574(7778):409-412. DOI:10.1038/s41586-019-1610-8 . |
| [83] | TAVERNER A M, YANG L, BARILE Z J, et al. Adaptive substitutions underlying cardiac glycoside insensitivity in insects exhibit epistasis in vivo[J]. eLife, 2019, 8: e48224. DOI:10.7554/eLife.48224 . |
| [1] | 谭贺, 杨晓辉, 张大秀, 王贵成. 不同发育时期小鼠代谢笼实验的最佳适应期探讨[J]. 实验动物与比较医学, 2024, 44(5): 502-510. |
| [2] | 李慧. 语言的比较生物学研究[J]. 实验动物与比较医学, 2022, 42(3): 255-261. |
| [3] | 尹媛, 陆璐, 王蕴, 杨景, 付鹤玲. 小鼠生活笼实验引入适应期的必要性研究[J]. 实验动物与比较医学, 2021, 41(4): 358-362. |
| [4] | 肖邦, 赵善民, 林丽芳, 王运慧, 宫晨, 余琛琳, 张璐, 汤球, 孙伟, 崔淑芳. 裸鼹鼠不同组织中低氧相关基因的表达[J]. 实验动物与比较医学, 2014, 34(5): 400-404. |
| [5] | 王希龙, 闵凡贵, 潘金春, 袁文, 陈瑞爱, 王凤国. 实验用五指山小型猪生长发育测定[J]. 实验动物与比较医学, 2011, 31(5): 352-355. |
| [6] | 陈克彪, 王志农, 王军, 黄盛东, 徐志云, 张宝仁. 山羊心肌缺血预适应实验模型的建立及其影响因素[J]. 实验动物与比较医学, 2005, 25(1): 30-33. |
| [7] | 连林生1,王鹤云1,徐家珍1,曾超登1,胡伟祥2,李光书2,赵毅2. 版纳微型猪的生物学特性[J]. 实验动物与比较医学, 1993, 13(4): 185-191. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

