
Laboratory Animal and Comparative Medicine ›› 2025, Vol. 45 ›› Issue (6): 762-772.DOI: 10.12300/j.issn.1674-5817.2025.111
• Invertebrate Laboratory Animals: Ant • Previous Articles Next Articles
Received:2025-07-05
Revised:2025-08-16
Online:2025-12-25
Published:2025-12-19
Contact:
SHENG Lihong
CLC Number:
SHENG Lihong. Harpegnathos saltator : A Model Insect for Decoding Plasticity of Social Behavior and Aging[J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 762-772. DOI: 10.12300/j.issn.1674-5817.2025.111.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2025.111
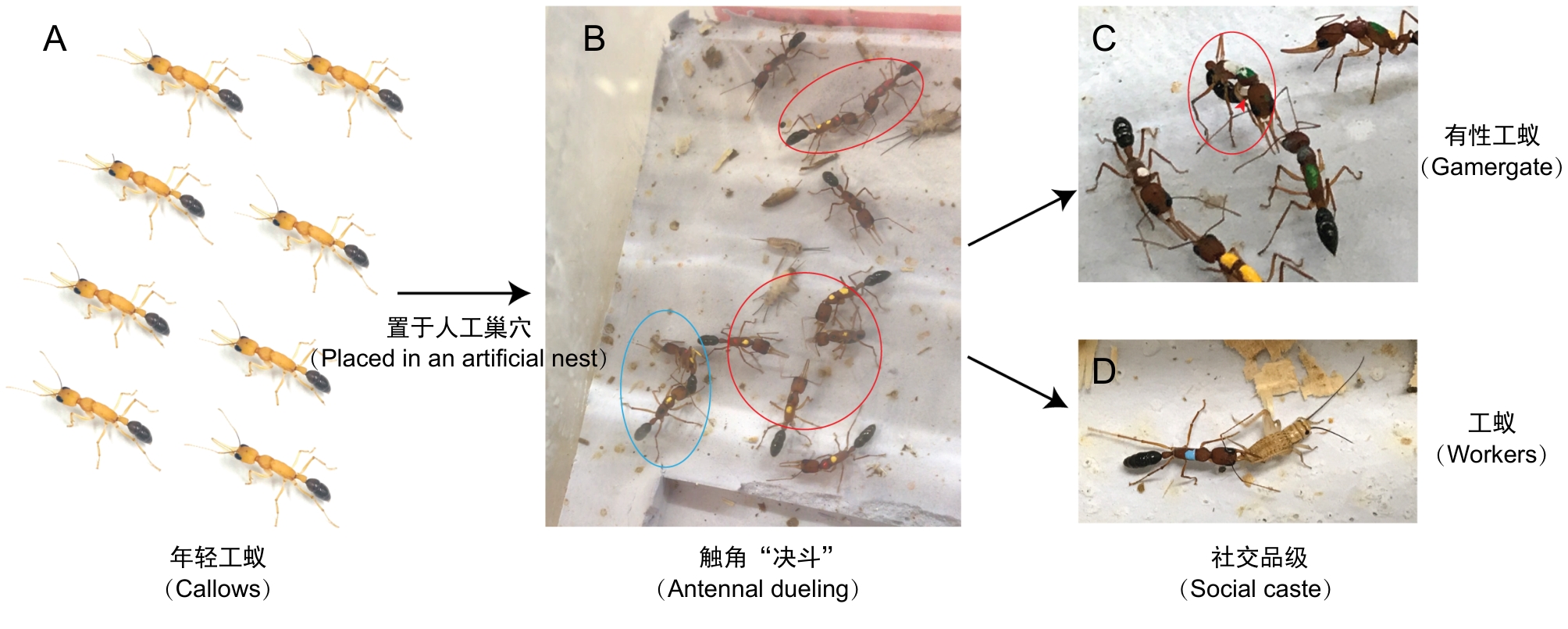
Figure 2 Establishment of a research model for caste transition in Harpegnathos saltator under laboratory conditionsNote: A, Callows; B, Caste transition, individuals marked with red circles are exhibiting antennal dueling behavior, and those marked with blue circles are performing "policing" behavior, which typically occurs when an isolated gamergate is reintroduced into the host colony; C, Egg-laying gamergate (red circle), with the red arrow indicating white egg; D, Foraging workers.
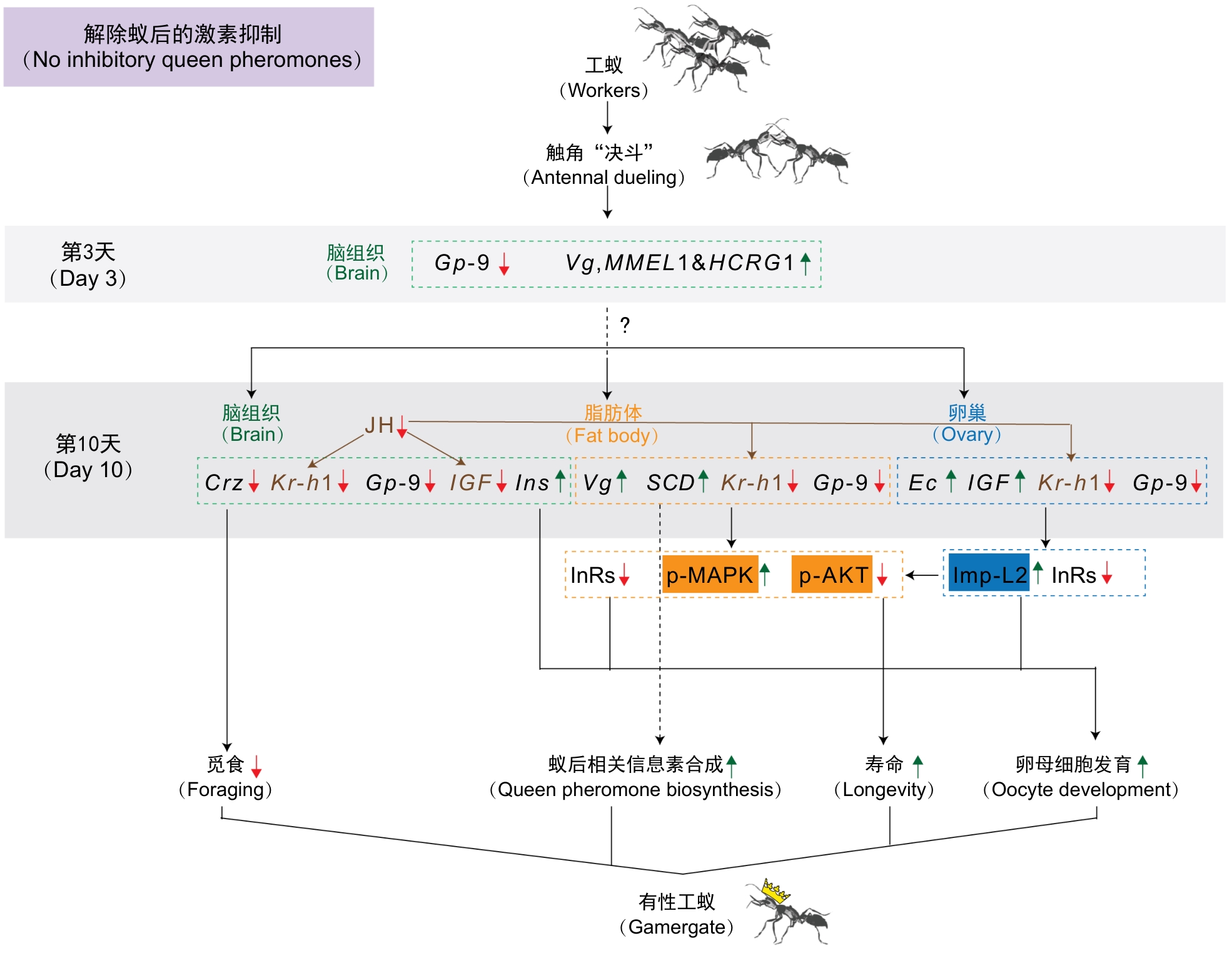
Figure 3 Schematic diagram of molecular changes related to caste transition in Harpegnathos saltator (modified from reference[14])Note: The absence of queen inhibitory pheromones relieves the suppression on antennal dueling behavior among workers and triggers a cascade of molecular changes across multiple tissues. Hypothetical model: initial gene expression changes in the destined gamergates brain, such as down-regulation of Gp-9 and up-regulation of vitellogenin, trigger a cascade of gene expression changes in the brain, fat body, and ovary that promote queen-like phenotypes, such as queen pheromone biosynthesis and oocyte development, and suppress worker-like phenotypes, such as foraging, while extending longevity. Ins is the homologous gene of human insulin, whereas IGF is the homologous gene of human IGF 1. Ins was previously known as Ilp1 and IGF as Ilp2 in H. saltator[9,15], while the homologous gene of Ins in honeybees and clonal raider ants was called Ilp2 [16-18]. Red arrows indicate downregulated molecules, green arrows indicate upregulated molecules in dueling ants. Solid lines represent confirmed molecule functions, while dashed lines represent hypothetical regulatory relationships.
| [1] | PYENSON B, ALBIN-BROOKS C, BURHYTE C, et al. Worker-like behavioral and physiological phenotype in queens with removed wings in a ponerine ant[J]. J Exp Biol, 2022, 225(18): jeb243684. DOI:10.1242/jeb.243684 . |
| [2] | PENICK C A, PRAGER S S, LIEBIG J. Juvenile hormone induces queen development in late-stage larvae of the ant Harpegnathos saltator [J]. J Insect Physiol, 2012, 58(12):1643-1649. DOI:10.1016/j.jinsphys.2012.10.004 . |
| [3] | HAIGHT K L. Patterns of venom production and temporal polyethism in workers of Jerdon's jumping ant, Harpegnathos saltator [J]. J Insect Physiol, 2012, 58(12):1568-1574. DOI:10.1016/j.jinsphys.2012.09.011 . |
| [4] | LIEBIG J, PEETERS C, OLDHAM N J, et al. Are variations in cuticular hydrocarbons of queens and workers a reliable signal of fertility in the ant Harpegnathos saltator?[J]. Proc Natl Acad Sci USA, 2000, 97(8):4124-4131. DOI: 10.1073/pnas.97.8.4124 . |
| [5] | GHANINIA M, HAIGHT K, BERGER S L, et al. Chemosensory sensitivity reflects reproductive status in the ant Harpegnathos saltator [J]. Sci Rep, 2017, 7(1):3732. DOI: 10.1038/s41598-017-03964-7 . |
| [6] | GELLERT H R, HALLEY D C, SIEB Z J, et al. Microstructures at the distal tip of ant chemosensory sensilla[J]. Sci Rep, 2022, 12(1):19328. DOI: 10.1038/s41598-022-21507-7 . |
| [7] | OPACHALOEMPHAN C, CARMONA-ALDANA F, YAN H. Caste transition and reversion in Harpegnathos saltator ant colonies[J]. Bio Protoc, 2023, 13(16): e4770. DOI: 10.21769/BioProtoc.4770 . |
| [8] | PENICK C A, GHANINIA M, HAIGHT K L, et al. Reversible plasticity in brain size, behaviour and physiology characterizes caste transitions in a socially flexible ant (Harpegnathos saltator)[J]. Proc Biol Sci, 2021, 288(1948):20210141. DOI: 10.1098/rspb.2021.0141 . |
| [9] | BONASIO R, ZHANG G J, YE C Y, et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator [J]. Science, 2010, 329(5995):1068-1071. DOI: 10.1126/science.1192428 . |
| [10] | SHIELDS E J, SHENG L H, WEINER A K, et al. High-quality genome assemblies reveal long non-coding RNAs expressed in ant brains[J]. Cell Rep, 2018, 23(10):3078-3090. DOI: 10.1016/j.celrep.2018.05.014 . |
| [11] | LIU A K, WANG Y, DANG C W, et al. A genome-wide identification and analysis of the basic helix-loop-helix transcription factors in the ponerine ant, Harpegnathos saltator [J]. BMC Evol Biol, 2012, 12:165. DOI:10.1186/1471-2148-12-165 . |
| [12] | BONASIO R, LI Q Y, LIAN J M, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator [J]. Curr Biol, 2012, 22(19):1755-1764. DOI: 10.1016/j.cub.2012.07.042 . |
| [13] | SHIELDS E J, SORIDA M, SHENG L H, et al. Genome annotation with long RNA reads reveals new patterns of gene expression and improves single-cell analyses in an ant brain[J]. BMC Biol, 2021, 19(1):254. DOI: 10.1186/s12915-021-01188-w . |
| [14] | OPACHALOEMPHAN C, MANCINI G, KONSTANTINIDES N, et al. Early behavioral and molecular events leading to caste switching in the ant Harpegnathos [J]. Genes Dev, 2021, 35(5-6):410-424. DOI: 10.1101/gad.343699.120 . |
| [15] | GOSPOCIC J, SHIELDS E J, GLASTAD K M, et al. The neuropeptide corazonin controls social behavior and caste identity in ants[J]. Cell, 2017, 170(4):748-759.e12. DOI: 10.1016/j.cell.2017.07.014 . |
| [16] | WANG Y, AZEVEDO S V, HARTFELDER K, et al. Insulin-like peptides (AmILP1 and AmILP2) differentially affect female caste development in the honey bee (Apis mellifera L.)[J]. J Exp Biol, 2013, 216(Pt 23):4347-4357. DOI: 10.1242/jeb.085779 . |
| [17] | CORONA M, VELARDE R A, REMOLINA S, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity[J]. Proc Natl Acad Sci USA, 2007, 104(17):7128-7133. DOI: 10.1073/pnas.0701909104 . |
| [18] | CHANDRA V, FETTER-PRUNEDA I, OXLEY P R, et al. Social regulation of insulin signaling and the evolution of eusociality in ants[J]. Science, 2018, 361(6400):398-402. DOI: 10.1126/science.aar5723 . |
| [19] | PENICK C A, BRENT C S, DOLEZAL K, et al. Neurohormonal changes associated with ritualized combat and the formation of a reproductive hierarchy in the ant Harpegnathos saltator [J]. J Exp Biol, 2014, 217(Pt 9):1496-1503. DOI: 10.1242/jeb.098301 . |
| [20] | WU Q, BROWN M R. Signaling and function of insulin-like peptides in insects[J]. Annu Rev Entomol, 2006, 51:1-24. DOI: 10.1146/annurev.ento.51.110104.151011 . |
| [21] | YAN H, OPACHALOEMPHAN C, CARMONA-ALDANA F, et al. Insulin signaling in the long-lived reproductive caste of ants[J]. Science, 2022, 377(6610):1092-1099. DOI: 10.1126/science.abm8767 . |
| [22] | LIBBRECHT R, OXLEY P R, KRONAUER D J C. Clonal raider ant brain transcriptomics identifies candidate molecular mechanisms for reproductive division of labor[J]. BMC Biol, 2018, 16(1):89. DOI: 10.1186/s12915-018-0558-8 . |
| [23] | PENICK C A, LIEBIG J, BRENT C S. Reproduction, dominance, and caste: endocrine profiles of queens and workers of the ant Harpegnathos saltator [J]. J Comp Physiol A Neuroethol Sens Neural Behav Physiol, 2011, 197(11):1063-1071. DOI: 10.1007/s00359-011-0667-0 . |
| [24] | BRENT C, PEETERS C, DIETEMANN V, et al. Hormonal correlates of reproductive status in the queenless ponerine ant, Streblognathus peetersi [J]. J Comp Physiol A Neuroethol Sens Neural Behav Physiol, 2006, 192(3):315-320. DOI: 10.1007/s00359-005-0065-6 . |
| [25] | BLOCH G, HEFETZ A, HARTFELDER K. Ecdysteroid titer, ovary status, and dominance in adult worker and queen bumble bees (Bombus terrestris)[J]. J Insect Physiol, 2000, 46(6):1033-1040. DOI: 10.1016/s0022-1910(99)00214-0 . |
| [26] | RACHINSKY A, STRAMBI C, STRAMBI A, et al. Caste and metamorphosis: hemolymph titers of juvenile hormone and ecdysteroids in last instar honeybee larvae[J]. Gen Comp Endocrinol, 1990, 79(1):31-38. DOI: 10.1016/0016-6480(90)90085-z . |
| [27] | GOSPOCIC J, GLASTAD K M, SHENG L H, et al. Kr-h1 maintains distinct caste-specific neurotranscriptomes in response to socially regulated hormones[J]. Cell, 2021, 184(23):5807-5823.e14. DOI: 10.1016/j.cell.2021.10.006 . |
| [28] | GLASTAD K M, ROESSLER J, GOSPOCIC J, et al. Long ant life span is maintained by a unique heat shock factor[J]. Genes Dev, 2023, 37(9-10):398-417. DOI: 10.1101/gad.350250.122 . |
| [29] | LÓPEZ-OTÍN C, BLASCO M A, PARTRIDGE L, et al. Hallmarks of aging: an expanding universe[J]. Cell, 2023, 186(2):243-278. DOI: 10.1016/j.cell.2022.11.001 . |
| [30] | SAHIN E, DEPINHO R A. Linking functional decline of telomeres, mitochondria and stem cells during ageing[J]. Nature, 2010, 464(7288):520-528. DOI: 10.1038/nature08982 . |
| [31] | SCHNEIDER S A, SCHRADER C, WAGNER A E, et al. Stress resistance and longevity are not directly linked to levels of enzymatic antioxidants in the ponerine ant Harpegnathos saltator [J]. PLoS One, 2011, 6(1): e14601. DOI: 10.1371/journal.pone.0014601 . |
| [32] | YAMAMOTO R, PALMER M, KOSKI H, et al. Aging modulated by the Drosophila insulin receptor through distinct structure-defined mechanisms[J]. Genetics, 2021, 217(2): iyaa037. DOI: 10.1093/genetics/iyaa037 . |
| [33] | LI W J, WANG C W, TAO L, et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans [J]. Nat Commun, 2021, 12(1):4568. DOI: 10.1038/s41467-021-24816-z . |
| [34] | MATHEW R, BHADRA M PAL, BHADRA U. Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species[J]. Biogerontology, 2017, 18(1):35-53. DOI: 10.1007/s10522-016-9670-8 . |
| [35] | COHEN E, PAULSSON J F, BLINDER P, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice[J]. Cell, 2009, 139(6):1157-1169. DOI: 10.1016/j.cell.2009.11.014 . |
| [36] | VIZUETA J, XIONG Z J, DING G, et al. Adaptive radiation and social evolution of the ants[J]. Cell, 2025, 188(18):4828-4848.e25. DOI: 10.1016/j.cell.2025.05.030 . |
| [37] | BONASIO R. Emerging topics in epigenetics: ants, brains, and noncoding RNAs[J]. Ann N Y Acad Sci, 2012, 1260:14-23. DOI: 10.1111/j.1749-6632.2011.06363.x . |
| [38] | SATO K, PELLEGRINO M, NAKAGAWA T, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels[J]. Nature, 2008, 452(7190):1002-1006. DOI: 10.1038/nature06850 . |
| [39] | SIMOLA D F, GRAHAM R J, BRADY C M, et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus [J]. Science, 2016, 351(6268): aac6633. DOI: 10.1126/science.aac6633 . |
| [40] | LINDQUIST S. The heat-shock response[J]. Annu Rev Biochem, 1986, 55:1151-1191. DOI: 10.1146/annurev.bi.55. 070186.005443 . |
| [41] | ÖSTLING P, BJÖRK J K, ROOS-MATTJUS P, et al. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1[J]. J Biol Chem, 2007, 282(10):7077-7086. DOI:10.1074/jbc.M607556200 . |
| [42] | SHENG L H, SHIELDS E J, GOSPOCIC J, et al. Social reprogramming in ants induces longevity-associated glia remodeling[J]. Sci Adv, 2020, 6(34): eaba9869. DOI:10.1126/sciadv.aba9869 . |
| [43] | SHENG L H, SHIELDS E J, GOSPOCIC J, et al. Ensheathing glia promote increased lifespan and healthy brain aging[J]. Aging Cell, 2023, 22(5): e13803. DOI:10.1111/acel.13803 . |
| [44] | FRAKES A E, METCALF M G, TRONNES S U, et al. Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans [J]. Science, 2020, 367(6476):436-440. DOI: 10.1126/science.aaz6896 . |
| [45] | KOMIYAMA T, LUO L Q. Development of wiring specificity in the olfactory system[J]. Curr Opin Neurobiol, 2006, 16(1):67-73. DOI: 10.1016/j.conb.2005.12.002 . |
| [46] | YAN H, JAFARI S, PASK G, et al. Evolution, developmental expression and function of odorant receptors in insects[J]. J Exp Biol, 2020, 223(Pt Suppl 1): jeb208215. DOI: 10.1242/jeb.208215 . |
| [47] | ROBERTSON H M. Molecular evolution of the major arthropod chemoreceptor gene families[J]. Annu Rev Entomol, 2019, 64:227-242. DOI: 10.1146/annurev-ento-020117-043322 . |
| [48] | RENGARAJAN S, HALLEM E A. Olfactory circuits and behaviors of nematodes[J]. Curr Opin Neurobiol, 2016, 41:136-148. DOI: 10.1016/j.conb.2016.09.002 . |
| [49] | IMAI T, SAKANO H. Odorant receptor-mediated signaling in the mouse[J]. Curr Opin Neurobiol, 2008, 18(3):251-260. DOI: 10.1016/j.conb.2008.07.009 . |
| [50] | BENTON R. On the ORigin of smell: odorant receptors in insects[J]. Cell Mol Life Sci, 2006, 63(14):1579-1585. DOI: 10.1007/s00018-006-6130-7 . |
| [51] | BUTTERWICK J A, DEL MÁRMOL J, KIM K H, et al. Cryo-EM structure of the insect olfactory receptor Orco[J]. Nature, 2018, 560(7719):447-452. DOI: 10.1038/s41586-018-0420-8 . |
| [52] | WICHER D, SCHÄFER R, BAUERNFEIND R, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels[J]. Nature, 2008, 452(7190):1007-1011. DOI: 10.1038/nature06861 . |
| [53] | BRAND P, ROBERTSON H M, LIN W, et al. The origin of the odorant receptor gene family in insects[J]. eLife, 2018, 7: e38340. DOI: 10.7554/eLife.38340 . |
| [54] | MIER P, FONTAINE J F, STOLDT M, et al. Annotation and analysis of 3902 odorant receptor protein sequences from 21 insect species provide insights into the evolution of odorant receptor gene families in solitary and social insects[J]. Genes (Basel), 2022, 13(5):919. DOI: 10.3390/genes13050919 . |
| [55] | ENGSONTIA P, SANGKET U, ROBERTSON H M, et al. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors[J]. BMC Res Notes, 2015, 8:380. DOI: 10.1186/s13104-015-1371-x . |
| [56] | ZHOU X F, SLONE J D, ROKAS A, et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding[J]. PLoS Genet, 2012, 8(8): e1002930. DOI: 10.1371/journal.pgen.1002930 . |
| [57] | YAN H, OPACHALOEMPHAN C, MANCINI G, et al. An engineered orco mutation produces aberrant social behavior and defective neural development in ants[J]. Cell, 2017, 170(4):736-747.e9. DOI: 10.1016/j.cell.2017.06.051 . |
| [58] | TRIBLE W, OLIVOS-CISNEROS L, MCKENZIE S K, et al. Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants[J]. Cell, 2017, 170(4):727-735.e10. DOI: 10.1016/j.cell.2017.07.001 . |
| [59] | SIERIEBRIENNIKOV B, SIEBER K R, KOLUMBA O, et al. Orco-dependent survival of odorant receptor neurons in ants[J]. Sci Adv, 2024, 10(23): eadk9000. DOI: 10.1126/sciadv.adk9000 . |
| [60] | PASK G M, SLONE J D, MILLAR J G, et al. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones[J]. Nat Commun, 2017, 8(1):297. DOI: 10.1038/s41467-017-00099-1 . |
| [61] | SLONE J D, PASK G M, FERGUSON S T, et al. Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator [J]. Proc Natl Acad Sci USA, 2017, 114(32):8586-8591. DOI: 10.1073/pnas.1704647114 . |
| [62] | LI Q Y, WANG M Y, ZHANG P, et al. A single-cell transcriptomic atlas tracking the neural basis of division of labour in an ant superorganism[J]. Nat Ecol Evol, 2022, 6(8):1191-1204. DOI: 10.1038/s41559-022-01784-1 . |
| [63] | JU L Y, GLASTAD K M, SHENG L H, et al. Hormonal gatekeeping via the blood-brain barrier governs caste-specific behavior in ants[J]. Cell, 2023, 186(20):4289-4309.e23. DOI: 10.1016/j.cell.2023.08.002 . |
| [64] | MCKENZIE S K, KRONAUER D J C. The genomic architecture and molecular evolution of ant odorant receptors[J]. Genome Res, 2018, 28(11):1757-1765. DOI: 10.1101/gr.237123.118 . |
| [65] | GAO Q H, XIONG Z J, LARSEN R S, et al. High-quality chromosome-level genome assembly and full-length transcriptome analysis of the Pharaoh ant Monomorium pharaonis [J]. Gigascience, 2020, 9(12): giaa143. DOI: 10.1093/gigascience/giaa143 . |
| [66] | WURM Y, WANG J, RIBA-GROGNUZ O, et al. The genome of the fire ant Solenopsis invicta [J]. Proc Natl Acad Sci USA, 2011, 108(14):5679-5684. DOI: 10.1073/pnas.1009690108 . |
| [67] | WANG M Y, LIU Y, WEN T G, et al. Chromatin accessibility and transcriptome landscapes of Monomorium pharaonis brain[J]. Sci Data, 2020, 7(1):217. DOI: 10.1038/s41597-020-0556-x . |
| [68] | JONES B M, WAUGH A H, CATTO M A, et al. The fire ant social chromosome exerts a major influence on genome regulation[J]. Mol Biol Evol, 2025, 42(6): msaf112. DOI: 10.1093/molbev/msaf112 . |
| [69] | GLASTAD K M, JU L Y, BERGER S L. Tramtrack acts during late pupal development to direct ant caste identity[J]. PLoS Genet, 2021, 17(9): e1009801. DOI: 10.1371/journal.pgen. 1009801 . |
| [70] | GLASTAD K M, GRAHAM R J, JU L Y, et al. Epigenetic regulator CoREST controls social behavior in ants[J]. Mol Cell, 2020, 77(2):338-351.e6. DOI: 10.1016/j.molcel.2019.10.012 . |
| [71] | SIEBER K, SAAR M, OPACHALOEMPHAN C, et al. Embryo injections for CRISPR-mediated mutagenesis in the ant Harpegnathos saltator [J]. J Vis Exp, 2021(168):10.3791/61930. DOI: 10.3791/61930 . |
| [72] | IVASYK I, OLIVOS-CISNEROS L, VALDÉS-RODRÍGUEZ S, et al. DNMT1 mutant ants develop normally but have disrupted oogenesis[J]. Nat Commun, 2023, 14(1):2201. DOI: 10.1038/s41467-023-37945-4 . |
| [73] | SIERIEBRIENNIKOV B, REINBERG D, DESPLAN C. A molecular toolkit for superorganisms[J]. Trends Genet, 2021, 37(9):846-859. DOI: 10.1016/j.tig.2021.05.005 . |
| [74] | HART T, FRANK D D, LOPES L E, et al. Sparse and stereotyped encoding implicates a core Glomerulus for ant alarm behavior[J]. Cell, 2023, 186(14):3079-3094.e17. DOI: 10.1016/j.cell.2023.05.025 . |
| [75] | ZHOU K C, HARFOUCHE M, COOKE C L, et al. Parallelized computational 3D video microscopy of freely moving organisms at multiple gigapixels per second[J]. Nat Photonics, 2023, 17(5):442-450. DOI: 10.1038/s41566-023-01171-7 . |
| [76] | KHILA A, ABOUHEIF E. In situ hybridization on ant ovaries and embryos[J]. Cold Spring Harb Protoc, 2009, 2009(7): pdb.prot5250. DOI: 10.1101/pdb.prot5250 . |
| [1] | SONG Mengjiao, SHEN Yidong. Approaches and Application Examples for Studying Mitochondrial Morphology and Function in Caenorhabditis elegans [J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 726-737. |
| [2] | XIAO Wenxian, LÜ Longbao. Research Progress on Human Ovarian Aging Using Non-Human Primates as Laboratory Animals [J]. Laboratory Animal and Comparative Medicine, 2025, 45(1): 47-54. |
| [3] | LIU Rongle, CHENG Hao, SHANG Fusheng, CHANG Shufu, XU Ping. Study on Cardiac Aging Phenotypes of SHJH hr Mice [J]. Laboratory Animal and Comparative Medicine, 2025, 45(1): 13-20. |
| [4] | ZHAO Lijuan, XIAO Chunlan, SHENG Yajie, LU Xi, ZHOU Zhengyu. Challenges and Development in Suzhou Laboratory Animal Industry Over the Past Five Decades [J]. Laboratory Animal and Comparative Medicine, 2024, 44(6): 645-653. |
| [5] | Ruiqi LI, Han DUAN, Luo GAN, Yuan ZHENG, Wen YANG. Advantages of Ciona intestinalis as a Model Organism and Its Applications [J]. Laboratory Animal and Comparative Medicine, 2024, 44(2): 162-179. |
| [6] | Yasheng DENG, Jiang LIN, Chiling GAN, Guanfeng ZENG, Jiayin HUANG, Huifang DENG, Yingxian MA, Siyin HAN. Literature Analysis of the Preparation Elements of Animal Models of Skin Photoaging and the Data of Subjects [J]. Laboratory Animal and Comparative Medicine, 2023, 43(4): 406-414. |
| [7] | Hui CHENG, Fei FANG, Jiahao SHI, Hua YANG, Mengjie ZHANG, Ping YANG, Jian FEI. H1 Linker Histone Gene Regulates Lifespan via Dietary Restriction Pathways in Caenorhabditis elegans [J]. Laboratory Animal and Comparative Medicine, 2023, 43(3): 271-281. |
| [8] | Danyang YIN, Yi HU, Rengfei SHI. Advances in Animal Aging Models [J]. Laboratory Animal and Comparative Medicine, 2023, 43(2): 156-162. |
| [9] | Huiqing TANG, Shufu CHANG, Zhifeng YU, Lei ZHANG, Xiaoqian TAN, Wei QU, Liang LI, Zhen QIAN, Jianzhong GU, Ping XU. Investigation on Biological Characteristics and Aging Phenotype of SHJH hr Mice [J]. Laboratory Animal and Comparative Medicine, 2023, 43(1): 44-52. |
| [10] | KONG Yue, GUO Yan. Construction and Evaluation of Skin Photoaging Mouse Model [J]. Laboratory Animal and Comparative Medicine, 2021, 41(2): 116-121. |
| [11] | CHEN Gaofeng, GAO Zhiling, LOU Weiwei, HUANG Lingying, JIN Shugen. Mice with Liver Fibrosis Based on Acoustic Radiation Force Impulse Imaging [J]. Laboratory Animal and Comparative Medicine, 2020, 40(5): 361-. |
| [12] | LEI Shan, LIU Qiang, HUANG Wei-jin, WANG You-chun. Influence of Strain, Gender and Hair Coat of Mice on Establishing Bioluminescent Imaging Pseudovirus Mouse Model [J]. Laboratory Animal and Comparative Medicine, 2019, 39(6): 423-428. |
| [13] | LU Jin, YAN Guo-feng, ZHOU Jin, XIAO Zhong-biao, ZHU Yin-qiu, ZHU Lian, WANG Jing, LI Yao, ZUO Yong. Comparison of Traditional Ultrasound with Speckle Tracking Imaging Technique by Myocardial Infarction Model in Mice [J]. Laboratory Animal and Comparative Medicine, 2018, 38(5): 356-364. |
| [14] | WANG Qin-zhou, ZHANG Cheng, LI Li, TANG Deng-xu, ZHANG Cai-qin, SHI Chang-hong. The Application of Heptamethine Cyanine Dye in Optical Imaging of Hepatocellular Carcinoma Transplantation Model [J]. Laboratory Animal and Comparative Medicine, 2018, 38(4): 250-254. |
| [15] | ZHOU Bin, SHENG Zhe-jin, FENG Chen-zhuo, LI Li-mei. A Melanoma Zebrafish Model for Real-time Imaging in vivo [J]. Laboratory Animal and Comparative Medicine, 2018, 38(1): 22-28. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

