
Laboratory Animal and Comparative Medicine ›› 2023, Vol. 43 ›› Issue (3): 271-281.DOI: 10.12300/j.issn.1674-5817.2022.183
• Model Animals and Animal Models • Previous Articles Next Articles
Hui CHENG1( ), Fei FANG1, Jiahao SHI1, Hua YANG1, Mengjie ZHANG1, Ping YANG2, Jian FEI1,2(
), Fei FANG1, Jiahao SHI1, Hua YANG1, Mengjie ZHANG1, Ping YANG2, Jian FEI1,2( )(
)( )
)
Received:2022-11-28
Revised:2023-02-15
Online:2023-06-25
Published:2023-06-25
Contact:
Jian FEI
CLC Number:
Hui CHENG,Fei FANG,Jiahao SHI,et al. H1 Linker Histone Gene Regulates Lifespan via Dietary Restriction Pathways in Caenorhabditis elegans[J]. Laboratory Animal and Comparative Medicine, 2023, 43(3): 271-281. DOI: 10.12300/j.issn.1674-5817.2022.183.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2022.183
基因 Gene | 引物序列 Sequences of primers |
|---|---|
| hil-1 | Forward: 5’-AATGGAACCGGAAGCAGCAA-3’ Reverse: 5’-TCCGCGAGTCTGTTCAATGT-3’ |
| daf-16 | Forward: 5’- GCTCCCTTCACTCGACACTT-3’ Reverse: 5’- TTCGATTGAGTTCGGGGACG-3’ |
| mtl-1 | Forward: 5’- GCTTGCAAGTGTGACTGCAA-3’ Reverse: 5’- TTGATGGGTCTTGTCTCCGC-3’ |
| ctl-1 | Forward: 5’- TACCGAGGAGGGTAACTGGG-3’ Reverse: 5’- AGTCTGTGGATTGCGCTTCA-3’ |
| act-4 | Forward: 5’- GCCACCGCTGCCTCCTCATC-3’ Reverse: 5’- CCGGCAGACTCCATACCCAAGAAG-3’ |
Table 1 Prime sequences for PCR
基因 Gene | 引物序列 Sequences of primers |
|---|---|
| hil-1 | Forward: 5’-AATGGAACCGGAAGCAGCAA-3’ Reverse: 5’-TCCGCGAGTCTGTTCAATGT-3’ |
| daf-16 | Forward: 5’- GCTCCCTTCACTCGACACTT-3’ Reverse: 5’- TTCGATTGAGTTCGGGGACG-3’ |
| mtl-1 | Forward: 5’- GCTTGCAAGTGTGACTGCAA-3’ Reverse: 5’- TTGATGGGTCTTGTCTCCGC-3’ |
| ctl-1 | Forward: 5’- TACCGAGGAGGGTAACTGGG-3’ Reverse: 5’- AGTCTGTGGATTGCGCTTCA-3’ |
| act-4 | Forward: 5’- GCCACCGCTGCCTCCTCATC-3’ Reverse: 5’- CCGGCAGACTCCATACCCAAGAAG-3’ |
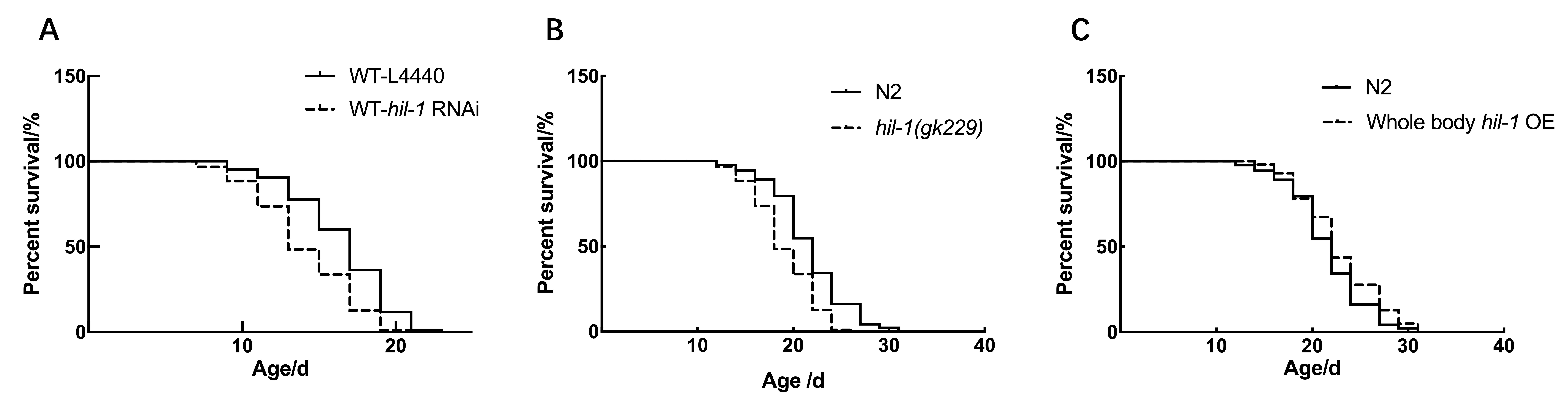
Figure 1 hil-1 is involved in regulation of the lifespan in C. elegansNote:A, Survival curves of wild-type N2 worms fed hil-1 RNA interference (RNAi) bacteria [compared with control group (fed L4440), P<0.001]; B, Survival curves of wild-type N2 worms and hil-1(gk229) mutants (compared with N2, P<0.001); C, Survival curves between wild-type N2 nematode and hil-1 systemic overexpression worms (compared with N2 nematode, P<0.05).
线虫种类 C. elegans strains | 线虫数量 Number | 平均寿命 Mean lifespan /d |
|---|---|---|
| N2 | 94 | 22±0.23 |
| hil-1(gk229) | 95 | 18±0.67*** |
| Whole body hil-1 OE | 101 | 22±0.22* |
| daf-2(e1370) | 101 | 47±0.46 |
| daf-2(e1370);hil-1(gk229) | 97 | 47±0.57 |
| daf-16(mu86) | 98 | 15±0.24 |
| daf-16(mu86);hil-1(gk229) | 82 | 14±0.26▲▲▲ |
| N2-L4440 | 85 | 17±0.23 |
| N2-hil-1 RNAi | 95 | 13±0.60△△△ |
| eat-2(ad465)-L4440 | 91 | 26±0.41 |
| eat-2(ad465)-hil-1 RNAi | 103 | 26±0.42 |
| NR350-L4440 | 90 | 16±0.56 |
| NR350-hil-1 RNAi | 95 | 16±0.57 |
| TU3311-L4440 | 101 | 13±0.28 |
| TU3311-hil-1 RNAi | 105 | 13±0.33 |
| NR222-L4440 | 105 | 22±0.27 |
| NR222-hil-1 RNAi | 91 | 17±0.34aaa |
| VP303-L4440 | 91 | 18±0.62 |
| VP303-hil-1 RNAi | 96 | 13±0.44bbb |
| AMJ345-L4440 | 99 | 15±0.21 |
| AMJ345-hil-1 RNAi | 99 | 15±0.23cc |
Table 2 Analysis of experimental data of lifespan in C. elegans
线虫种类 C. elegans strains | 线虫数量 Number | 平均寿命 Mean lifespan /d |
|---|---|---|
| N2 | 94 | 22±0.23 |
| hil-1(gk229) | 95 | 18±0.67*** |
| Whole body hil-1 OE | 101 | 22±0.22* |
| daf-2(e1370) | 101 | 47±0.46 |
| daf-2(e1370);hil-1(gk229) | 97 | 47±0.57 |
| daf-16(mu86) | 98 | 15±0.24 |
| daf-16(mu86);hil-1(gk229) | 82 | 14±0.26▲▲▲ |
| N2-L4440 | 85 | 17±0.23 |
| N2-hil-1 RNAi | 95 | 13±0.60△△△ |
| eat-2(ad465)-L4440 | 91 | 26±0.41 |
| eat-2(ad465)-hil-1 RNAi | 103 | 26±0.42 |
| NR350-L4440 | 90 | 16±0.56 |
| NR350-hil-1 RNAi | 95 | 16±0.57 |
| TU3311-L4440 | 101 | 13±0.28 |
| TU3311-hil-1 RNAi | 105 | 13±0.33 |
| NR222-L4440 | 105 | 22±0.27 |
| NR222-hil-1 RNAi | 91 | 17±0.34aaa |
| VP303-L4440 | 91 | 18±0.62 |
| VP303-hil-1 RNAi | 96 | 13±0.44bbb |
| AMJ345-L4440 | 99 | 15±0.21 |
| AMJ345-hil-1 RNAi | 99 | 15±0.23cc |
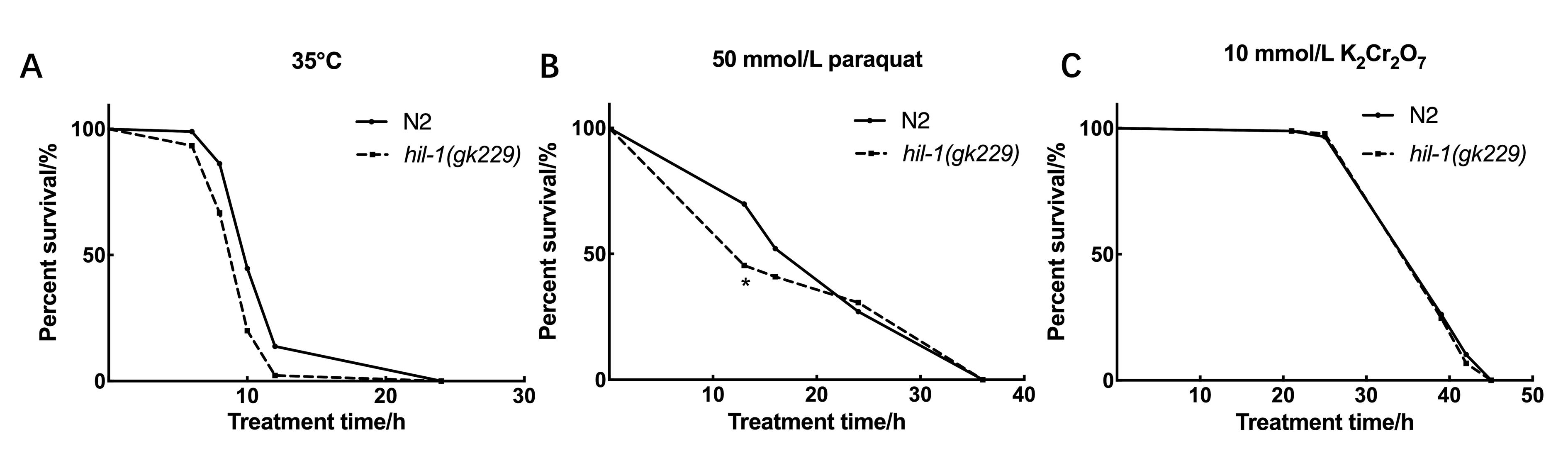
Figure 2 The deletion of hil-1 reduces the tolerance to heat shock and oxidative stress of C. elegansNote:A, Survival curves of wild-type N2 worms and hil-1(gk229) mutant worms transferred from 20 °C to 35 °C, P<0.001; B, Survival curves of wild-type N2 worms and hil-1(gk229) mutant worms exposed to 50 mmol/L paraquat, *P<0.05; C, Survival curves of wild-type N2 worms and hil-1(gk229) mutant worms exposed to 10 mmol/L K2Cr2O7, P>0.05.
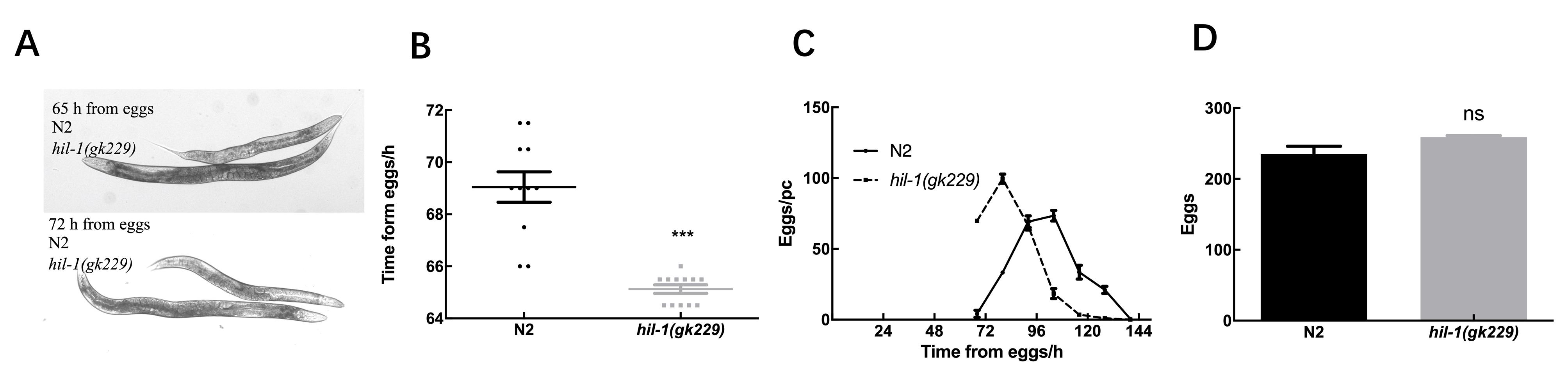
Figure 3 The deletion of hil-1 affects the development of C. elegansNote:A, Observing the morphology of eggs of wild-type N2 worms and hil-1(gk229) mutant worms at 65 h and 72 h of development under microscope, with magnification of 100X; B, The duration of wild-type N2 worms and hil-1(gk229) mutant worms from egg to the laying of the first egg, n =10 in each group, ***P<0.001; C, The average number of eggs production every 12 h from egg development in wild-type N2 worms and hil-1(gk229) mutant worms, n > 10 in each group; D, The total number of eggs in wild-type N2 worms and hil-1(gk229) mutant worms. n > 10 in each group, nsP>0.05.
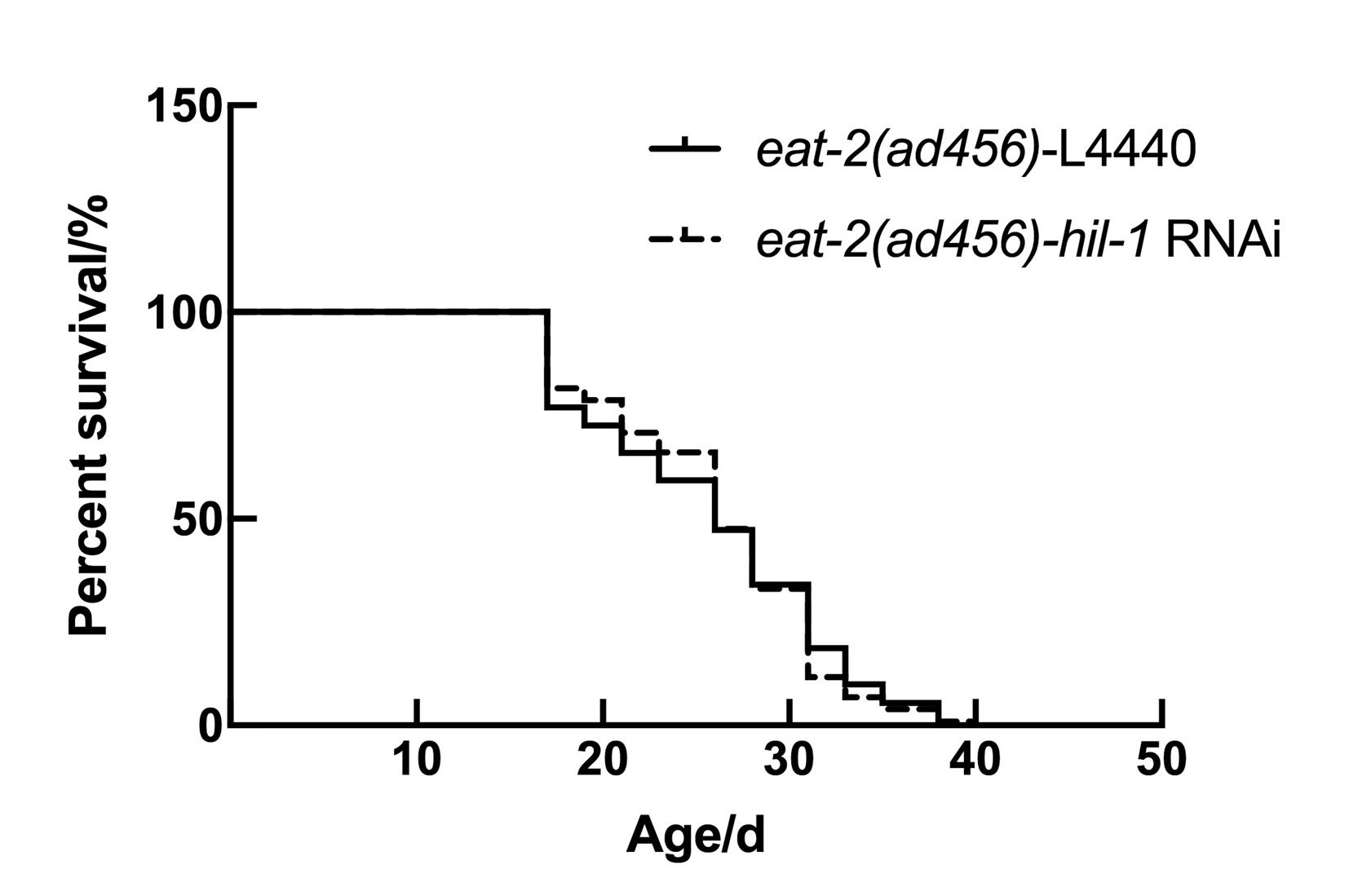
Figure 4 The knockdown of hil-1 has no significant effect on the lifespan of eat-2(ad465)Note: Survival curves of eat-2(ad465) mutants fed hil-1 RNA interference (RNAi) bacteria. Compared with control group (fed L4440), P>0.05.
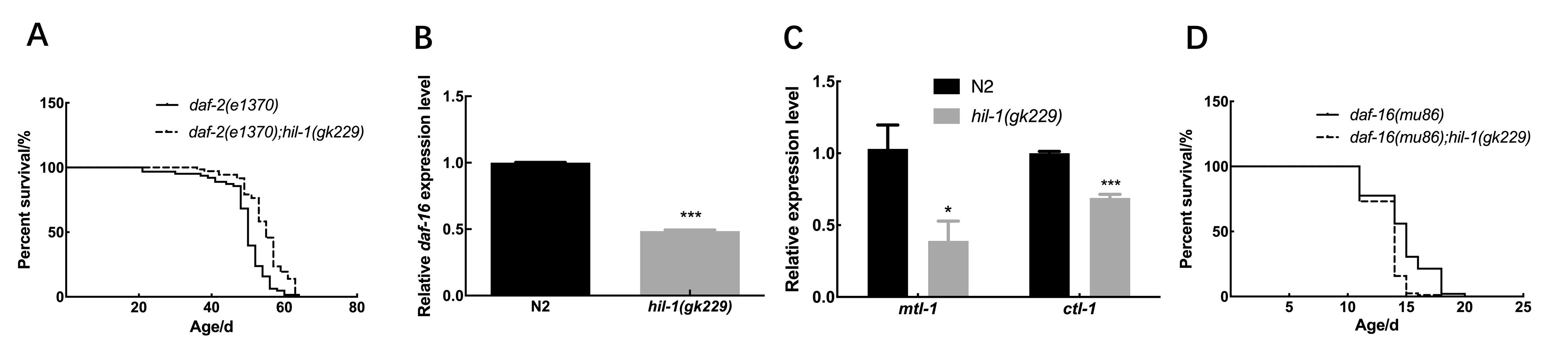
Figure 5 hil-1 is not completely dependent on insulin signaling pathway to regulate lifespan of C. elegansNote:A, Survival curves of daf-2(e1370) mutant worms and daf-2(e1370);hil-1(gk229) mutant worms, P>0.05; B, Analysis of the expression levels of daf-16 gene in wild-type N2 worms and hil-1(gk229) mutant worms(Compared with wild-type N2 worms, ***P<0.001); C, Analysis of the expression levels of daf-16 target genes mtl-1 and ctl-1 in wild-type N2 and hil-1(gk229) mutant worms(compared with wild-type N2 worms *P<0.05,***P<0.001); D, Survival curves of daf-16(mu86) mutant worms and daf-16 (mu86); hil-1(gk229) mutant worms, P<0.001.
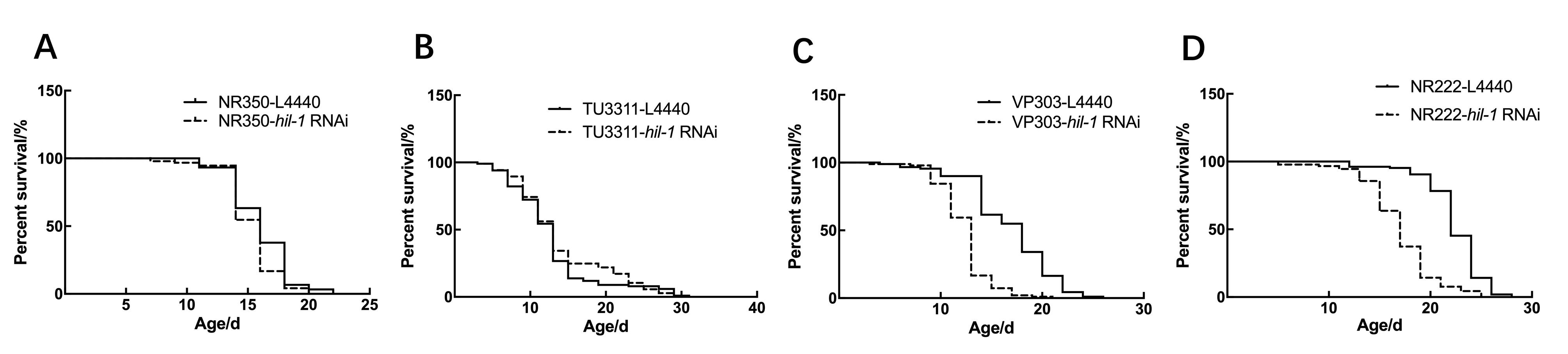
Figure 6 hil-1 acts in the hypodermis or intestine to regulate lifespanNote:A-D, Survival curves of worms with tissue-specific knockdown of hil-1in muscle(A), neuron(B), hypodermis(C) and intestine(D), respectively. Lifespan is shortened only in hypodermis-specific RNA interference (RNAi) or intestine-specific RNAi worms.
| 1 | LÓPEZ-OTÍN C, BLASCO M A, PARTRIDGE L, et al. The hallmarks of aging[J]. Cell, 2013, 153(6):1194-1217. DOI: 10.1016/j.cell.2013.05.039 . |
| 2 | NIGON V M, FÉLIX M A. History of research on C. elegans and other free-living nematodes as model organisms[J]. WormBook, 2017, 2017:1-84. DOI: 10.1895/wormbook.1.181.1 . |
| 3 | HONJOH S, YAMAMOTO T, UNO M, et al. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans [J]. Nature, 2009, 457(7230):726-730. DOI: 10.1038/nature07583 . |
| 4 | KENYON C, CHANG J, GENSCH E, et al. A C. elegans mutant that lives twice as long as wild type[J]. Nature, 1993, 366(6454):461-464. DOI: 10.1038/366461a0 . |
| 5 | VELLAI T, TAKACS-VELLAI K, ZHANG Y, et al. Genetics: influence of TOR kinase on lifespan in C. elegans [J]. Nature, 2003, 426(6967):620. |
| 6 | HARRISON D E, STRONG R, DAVE SHARP Z, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice[J]. Nature, 2009, 460(7253):392-395. DOI: 10.1038/nature08221 . |
| 7 | KAEBERLEIN M, POWERS R W 3rd, STEFFEN K K, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients[J]. Science, 2005, 310(5751):1193-1196. |
| 8 | KENYON C. The plasticity of aging: insights from long-lived mutants[J]. Cell, 2005, 120(4):449-460. |
| 9 | KAPAHI P, ZID B M, HARPER T, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway[J]. Curr Biol, 2004, 14(10):885-890. |
| 10 | NOLL M, KOROBERG R D. Action of micrococcal nuclease on chromatin and the location of histone H1[J]. J Mol Biol, 1977, 109(3):393-404. DOI: 10.1016/S0022-2836(77)80019-3 . |
| 11 | ALLAN J, HARTMAN P G, CRANE-ROBINSON C, et al. The structure of histone H1 and its location in chromatin[J]. Nature, 1980, 288(5792):675-679. DOI: 10.1038/288675a0 . |
| 12 | HERGETH S P, SCHNEIDER R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle[J]. EMBO Rep, 2015, 16(11):1439-1453. DOI: 10.15252/embr.201540749 . |
| 13 | HAO F F, KALE S, DIMITROV S, et al. Unraveling linker histone interactions in nucleosomes[J]. Curr Opin Struct Biol, 2021, 71: S0959-S440X(21)00083-X. DOI: 10.1016/j.sbi.2021.06.001 . |
| 14 | BHAN S, MAY W, WARREN S L, et al. Global gene expression analysis reveals specific and redundant roles for H1 variants, H1c and H1(0), in gene expression regulation[J]. Gene, 2008, 414(1-2):10-18. DOI: 10.1016/j.gene.2008.01.025 . |
| 15 | DI LIEGRO C, SCHIERA G, DI LIEGRO I. H1.0 linker histone as an epigenetic regulator of cell proliferation and differentiation[J]. Genes, 2018, 9(6):310. DOI: 10.3390/genes9060310 . |
| 16 | 石锦平, 任长虹, 李媛, 等. 秀丽隐杆线虫双突变株系hif-1(ia04);glb-13(tm2825)的制备与鉴定[J]. 军事医学, 2011, 35(4):253-257. DOI:10.3969/j.issn.1674-9960.2011.04.004 . |
| SHI J P, REN C H, LI Y, et al. Construction and characterization of double mutant strain hif-1(ia04);glb-13(tm2825) in C.elegans [J]. Military Medical Sciences, 2011, 35(4):253-257. DOI:10.3969/j.issn.1674-9960.2011.04.004 . | |
| 17 | ZHAO Y, GILLIAT A F, ZIEHM M, et al. Two forms of death in ageing Caenorhabditis elegans [J]. Nat Commun, 2017, 8:15458. DOI: 10.1038/ncomms15458 . |
| 18 | YANG P, SUN R L, YAO M H, et al. A C-terminal truncated mutation of spr-3 gene extends lifespan in Caenorhabditis elegans [J]. Acta Biochim Biophys Sin (Shanghai), 2013, 45(7):540-548. DOI: 10.1093/abbs/gmt048 . |
| 19 | LARANCE M, POURKARIMI E, WANG B, et al. Global proteomics analysis of the response to starvation in C. elegans [J]. Mol Cell Proteomics, 2015, 14(7):1989-2001. DOI: 10.1074/mcp.M114.044289 . |
| 20 | JEDRUSIK M A, VOGT S, CLAUS P, et al. A novel linker histone-like protein is associated with cytoplasmic filaments in Caenorhabditis elegans [J]. J Cell Sci, 2002, 115(Pt 14):2881-2891. |
| 21 | HUANG C, XIONG C J, KORNFELD K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans [J]. Proc Natl Acad Sci USA, 2004, 101(21):8084-8089. |
| 22 | LEE S S, KENNEDY S, TOLONEN A C, et al. DAF-16 target genes that control C. elegans life-span and metabolism[J]. Science, 2003, 300(5619): 644-7. DOI: 10.1126/science.1083614 . |
| 23 | SUN X, CHEN W D, WANG Y D. DAF-16/FOXO transcription factor in aging and longevity [J]. Front Pharmacol, 2017, 8: 548. DOI: 10.3389/fphar.2017.00548 . |
| 24 | 尹丹阳, 胡怡, 史仍飞. 动物衰老模型的研究进展[J]. 实验动物与比较医学, 2023, 43(2): 156-162. DOI: 10.12300/j.issn.1674-5817.2022.094 . |
| YIN D Y, HU Y, SHI R F. Advances in animal aging models[J]. Lab Anim Comp Med, 2023, 43(2): 156-162. DOI: 10.12300/j.issn.1674-5817.2022.094 . | |
| 25 | HARSHMAN S W, YOUNG N L, PARTHUN M R, et al. H1 histones: current perspectives and challenges[J]. Nucleic Acids Res, 2013, 41(21):9593-9609. DOI: 10.1093/nar/gkt700 . |
| 26 | PAN C Y, FAN Y H. Role of H1 linker histones in mammalian development and stem cell differentiation[J]. Biochim Biophys Acta, 2016, 1859(3): S1874-S9399(15)00266-7. DOI: 10.1016/j.bbagrm.2015.12.002 . |
| 27 | SUN J, WEI H M, XU J, et al. Histone H1-mediated epigenetic regulation controls germline stem cell self-renewal by modulating H4K16 acetylation[J]. Nat Commun, 2015, 6:8856. DOI: 10.1038/ncomms9856 . |
| 28 | YANASE S, YASUDA K, ISHII N. Adaptive responses to oxidative damage in three mutants of Caenorhabditis elegans (age-1, mev-1 and daf-16) that affect life span[J]. Mech Ageing Dev, 2002, 123(12):1579-1587. |
| 29 | ANBALAGAN C, LAFAYETTE I, ANTONIOU-KOUROUNIOTI M, et al. Transgenic nematodes as biosensors for metal stress in soil pore water samples[J]. Ecotoxicology, 2012, 21(2):439-455. DOI: 10.1007/s10646-011-0804-0 . |
| 30 | MOILANEN L H, FUKUSHIGE T, FREEDMAN J H. Regulation of metallothionein gene transcription[J]. J Biol Chem, 1999, 274(42):29655-29665. DOI: 10.1074/jbc.274.42.29655 . |
| 31 | LIN C X, ZHANG X Y, SU Z X, et al. Carnosol improved lifespan and healthspan by promoting antioxidant capacity in Caenorhabditis elegans [J]. Oxid Med Cell Longev, 2019, 2019:5958043. DOI: 10.1155/2019/5958043 . |
| 32 | GREEN C L, LAMMING D W, FONTANA L. Molecular mechanisms of dietary restriction promoting health and longevity[J]. Nat Rev Mol Cell Biol, 2022, 23(1):56-73. DOI: 10.1038/s41580-021-00411-4 . |
| 33 | ZHANG Y P, ZHANG W H, ZHANG P, et al. Intestine-specific removal of DAF-2 nearly doubles lifespan in Caenorhabditis elegans with little fitness cost[J]. Nat Commun, 2022, 13(1):6339. DOI: 10.1038/s41467-022-33850-4 . |
| 34 | HAHM J H, JEONG C, NAM H G. Diet restriction-induced healthy aging is mediated through the immune signaling component ZIP-2 in Caenorhabditis elegans [J]. Aging Cell, 2019, 18(5): e12982. DOI: 10.1111/acel.12982 . |
| 35 | WEIR H J, YAO P, HUYNH F K, et al. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling[J]. Cell Metab, 2017, 26(6): S1550-S4131(17)30612-5. DOI: 10.1016/j.cmet.2017.09.024 . |
| 36 | CAI H H, RASULOVA M, VANDEMEULEBROUCKE L, et al. Life-span extension by axenic dietary restriction is independent of the mitochondrial unfolded protein response and mitohormesis in Caenorhabditis elegans [J]. J Gerontol A Biol Sci Med Sci, 2017, 72(10):1311-1318. DOI: 10.1093/gerona/glx013 . |
| 37 | SMITH-VIKOS T, DE LENCASTRE A, INUKAI S, et al. MicroRNAs mediate dietary-restriction-induced longevity through PHA-4/FOXA and SKN-1/Nrf transcription factors[J]. Curr Biol, 2014, 24(19): S0960-S9822(14)00991-9. DOI: 10.1016/j.cub.2014.08.013 . |
| [1] | XIAO Wenxian, LÜ Longbao. Research Progress on Human Ovarian Aging Using Non-Human Primates as Laboratory Animals [J]. Laboratory Animal and Comparative Medicine, 2025, 45(1): 47-54. |
| [2] | LIU Rongle, CHENG Hao, SHANG Fusheng, CHANG Shufu, XU Ping. Study on Cardiac Aging Phenotypes of SHJH hr Mice [J]. Laboratory Animal and Comparative Medicine, 2025, 45(1): 13-20. |
| [3] | ZHAO Lijuan, XIAO Chunlan, SHENG Yajie, LU Xi, ZHOU Zhengyu. Challenges and Development in Suzhou Laboratory Animal Industry Over the Past Five Decades [J]. Laboratory Animal and Comparative Medicine, 2024, 44(6): 645-653. |
| [4] | Yasheng DENG, Jiang LIN, Chiling GAN, Guanfeng ZENG, Jiayin HUANG, Huifang DENG, Yingxian MA, Siyin HAN. Literature Analysis of the Preparation Elements of Animal Models of Skin Photoaging and the Data of Subjects [J]. Laboratory Animal and Comparative Medicine, 2023, 43(4): 406-414. |
| [5] | Danyang YIN, Yi HU, Rengfei SHI. Advances in Animal Aging Models [J]. Laboratory Animal and Comparative Medicine, 2023, 43(2): 156-162. |
| [6] | Huiqing TANG, Shufu CHANG, Zhifeng YU, Lei ZHANG, Xiaoqian TAN, Wei QU, Liang LI, Zhen QIAN, Jianzhong GU, Ping XU. Investigation on Biological Characteristics and Aging Phenotype of SHJH hr Mice [J]. Laboratory Animal and Comparative Medicine, 2023, 43(1): 44-52. |
| [7] | KONG Yue, GUO Yan. Construction and Evaluation of Skin Photoaging Mouse Model [J]. Laboratory Animal and Comparative Medicine, 2021, 41(2): 116-121. |
| [8] | CHEN Gaofeng, GAO Zhiling, LOU Weiwei, HUANG Lingying, JIN Shugen. Mice with Liver Fibrosis Based on Acoustic Radiation Force Impulse Imaging [J]. Laboratory Animal and Comparative Medicine, 2020, 40(5): 361-. |
| [9] | LEI Shan, LIU Qiang, HUANG Wei-jin, WANG You-chun. Influence of Strain, Gender and Hair Coat of Mice on Establishing Bioluminescent Imaging Pseudovirus Mouse Model [J]. Laboratory Animal and Comparative Medicine, 2019, 39(6): 423-428. |
| [10] | LU Jin, YAN Guo-feng, ZHOU Jin, XIAO Zhong-biao, ZHU Yin-qiu, ZHU Lian, WANG Jing, LI Yao, ZUO Yong. Comparison of Traditional Ultrasound with Speckle Tracking Imaging Technique by Myocardial Infarction Model in Mice [J]. Laboratory Animal and Comparative Medicine, 2018, 38(5): 356-364. |
| [11] | WANG Qin-zhou, ZHANG Cheng, LI Li, TANG Deng-xu, ZHANG Cai-qin, SHI Chang-hong. The Application of Heptamethine Cyanine Dye in Optical Imaging of Hepatocellular Carcinoma Transplantation Model [J]. Laboratory Animal and Comparative Medicine, 2018, 38(4): 250-254. |
| [12] | ZHOU Bin, SHENG Zhe-jin, FENG Chen-zhuo, LI Li-mei. A Melanoma Zebrafish Model for Real-time Imaging in vivo [J]. Laboratory Animal and Comparative Medicine, 2018, 38(1): 22-28. |
| [13] | LIU Qian, LIU Song, Han Feng-feng, GUO Xue-jun. Establishment of Orthotopic Transplant Model of Human Lung Cancer and Observation of In vivo Biofluorescent Imaging in Nude Mice [J]. Laboratory Animal and Comparative Medicine, 2016, 36(4): 250-256. |
| [14] | ZHAO Yong-jiang, ZHU Miao-xin, YUAN Li-xin, SUN Lei, GENG Qin, LI Jing, YAO Ming, YAN Ming-xia. Dynamic Observation on In vivo Biofluorescent Imaging of Orthotopic Transplant Model of Human Colon Cancer in Nude Mice [J]. Laboratory Animal and Comparative Medicine, 2016, 36(3): 174-179. |
| [15] | XI Xiao-xia, XI Dong-bin, FAN Lin-lan, WEI Hu-lai. Anti-metastatic Actions of Realgar Nanoparticles on Hepatic and Pulmonary Metastasis of Murine Malignant Melanoma with in vivo Bioluminescence Imaging System [J]. Laboratory Animal and Comparative Medicine, 2015, 35(5): 345-350. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||