













实验动物与比较医学 ›› 2024, Vol. 44 ›› Issue (1): 62-73.DOI: 10.12300/j.issn.1674-5817.2023.079
收稿日期:2023-06-16
修回日期:2023-10-13
出版日期:2024-02-25
发布日期:2024-03-07
通讯作者:
孔 琪(1978—),男,博士,研究员,研究方向:比较医学。E-mail: kongqi@ cnilas.org。ORCID: 0000-0003-2867-7382作者简介:冯婷婷(1999—),女,硕士研究生,研究方向:比较医学。E-mail: fengtingting@ cnilas.org
基金资助:
Tingting FENG, Yitong LI, Yue WU, Jue WANG, Qi KONG( )(
)( )
)
Received:2023-06-16
Revised:2023-10-13
Published:2024-02-25
Online:2024-03-07
Contact:
KONG Qi (ORCID: 0000-0003-2867-7382), E-mail: kongqi@ cnilas.org摘要:
目的 通过系统梳理与分析新冠病毒感染相关的转录组数据,为研究新冠病毒感染过程中的生物学变化和致病机制提供更多的比较医学基础信息。 方法 按照检索策略,以COVID-19和SARS-CoV-2为检索词,从GEO、ArrayExpress、GEN三大转录组数据库收集获得2020年1月—2023年5月的新冠病毒感染转录组数据集,分析得到新冠病毒感染转录组的数据资源组成、分布和研究应用情况,并对数据分布进行可视化展示及关联分析。从临床医学和比较医学两个方面,围绕临床相关分子机制、生物标志物及相关免疫反应、治疗干预策略等方面,分析现有新冠病毒感染转录组数据的研究应用和局限性,阐述其研究价值和应用前景。 结果 共纳入新冠病毒感染转录组数据975组,在3个数据库中样本来源于人的数据集最多,分别占71.9%、77.9%和90.0%。除人以外,小鼠等是数据主要来源物种,呼吸系统和神经系统是数据分布排名在前的两个系统。在临床意义方面关联27个数据集。分析显示,基于转录组数据挖掘得到呼吸道损伤等相关分子机制,cfDNA等生物标志物可作为治疗靶点,以M1型巨噬细胞为代表的细胞变化和失调与新冠病毒感染严重程度相关。比较医学分析表明小鼠、仓鼠等均为新冠病毒易感动物,其中恒河猴和食蟹猴与人类的感染特征高度相似,仓鼠除了呼吸道症状还存在消化系统症状。新冠病毒能在各种易感动物呼吸系统器官、雪貂的肠道及水貂的耳部进行复制,产生肺炎、弥漫性肺损伤等不同程度的病理变化;基于免疫应答差异,可使用仓鼠进行中和抗体反应研究。 结论 目前新冠病毒感染转录组数据较多,但缺少比较转录组研究,可将转录组学与比较医学进一步结合,从而对新冠病毒感染的比较医学差异进行更深入的挖掘。
中图分类号:
冯婷婷, 李依桐, 吴玥, 王珏, 孔琪. 新冠病毒感染转录组学数据及比较医学分析[J]. 实验动物与比较医学, 2024, 44(1): 62-73.
Tingting FENG, Yitong LI, Yue WU, Jue WANG, Qi KONG. Transcriptome Data and Comparative Medical Analysis of COVID-19 Virus Infection[J]. Laboratory Animal and Comparative Medicine, 2024, 44(1): 62-73.
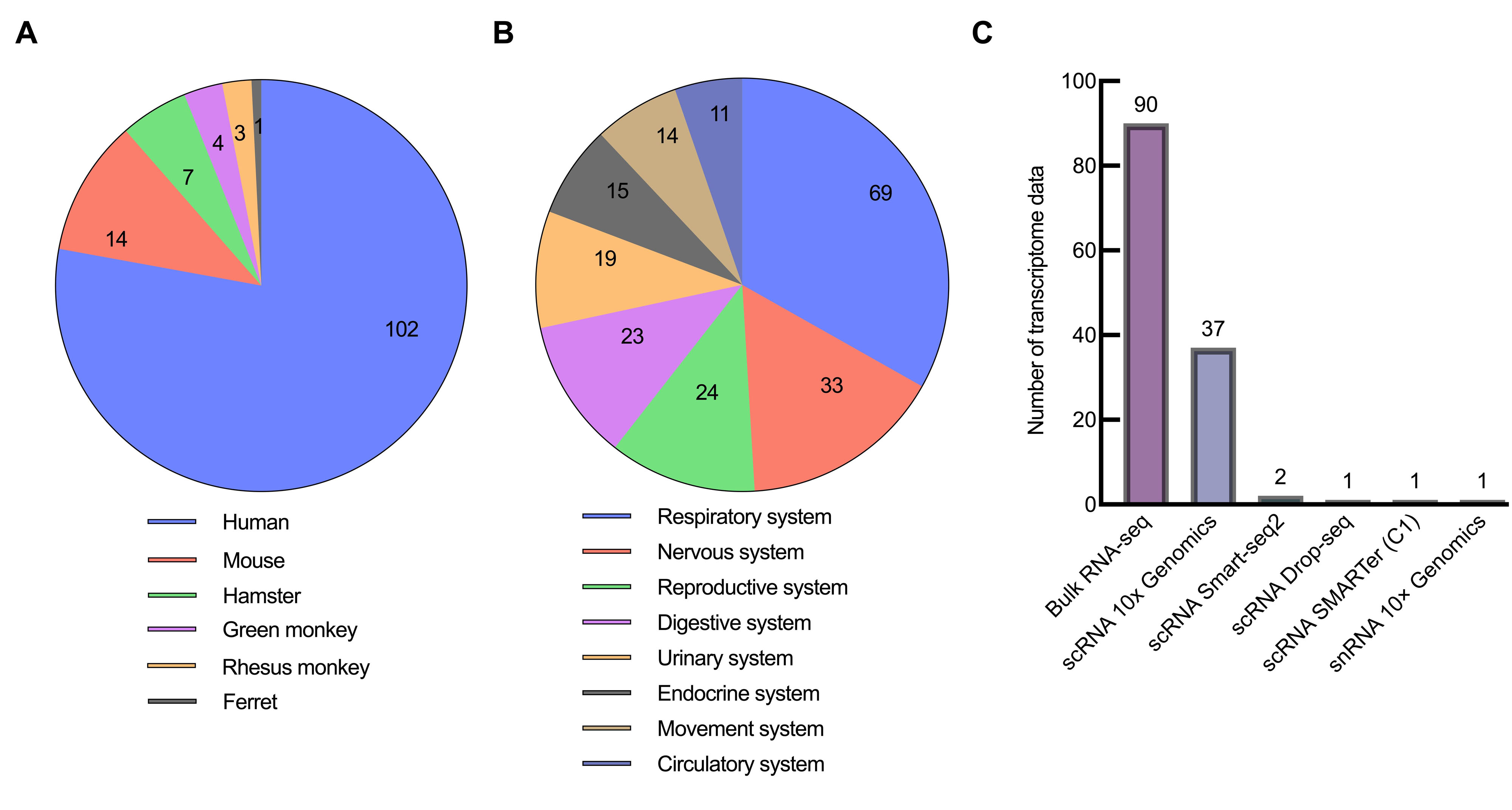
图3 GEN数据库中SARS-CoV-2感染转录组数据分类结果注:A,按研究物种分类;B,按感染系统分类;C,按测序方法分类。
Figure 3 Classification result of SARS-CoV-2 infection transcriptome data in GEN databaseNote:A, classified by species; B, classified by system; C, classified by sequencing methods.
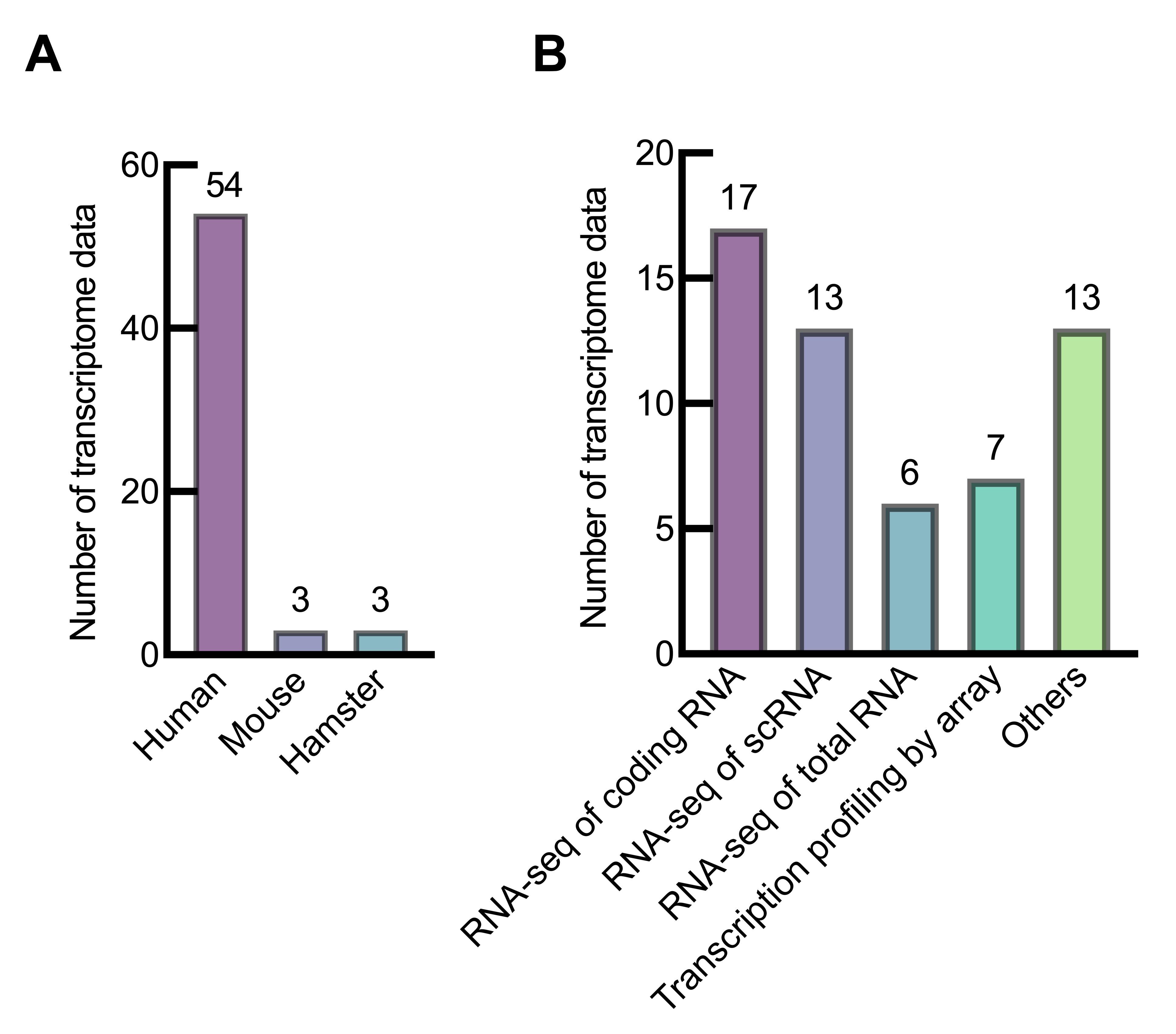
图4 ArrayExpress数据库中SARS-CoV-2感染转录组数据分类结果注:A,按物种分类;B,按具体方法分类。
Figure 4 Classification results of SARS-CoV-2 infection transcriptome data in the ArrayExpress databaseNote:A, classified by species; B, classified by specific methods.
样本来源 Sample origins | 组织/细胞 Tissues/cells | 发现 Finding | 数据集编号 Datasets' number |
|---|---|---|---|
人;雪貂 Human; ferrets | NHBE、A549、Calu3细胞,肺组织(人); 鼻腔冲洗液(雪貂) | 新冠病毒感染肺损伤相关机制 | GSE147507 |
人;雪貂 Human; ferrets | NHBE、A549、Calu3细胞(人);鼻腔冲洗 液(雪貂) | 应对新冠病毒感染时NHBE细胞上调MALAT1和NEAT1表达 | GSE147507 |
人;C57BL/6小鼠 Human; C57BL/6 mice | PBMC、肺泡灌洗液(人);淋巴结(小鼠) | 髓细胞、上皮细胞和T细胞之间的相互作用驱动组织损伤 | GSE149689; GSE155673; GSE145926; GSE149443 |
金黄仓鼠 Golden hamsters | 心脏 | 新冠病毒感染对心脏的损伤机制 | PRJNA884511 |
| 人Human | 肺、肝脏 | 新冠病毒感染肝损伤相关机制 | GSE150316 |
| 人Human | 肺泡灌洗液 | 严重新冠病毒感染患者的支气管肺泡灌洗液中富含促炎单核细胞衍生的巨噬细胞 | GSE145926 |
| 人Human | 肺泡灌洗液 | M1型巨噬细胞和重度新冠病毒感染患者之间的关系最高 | GSE145926 |
| 人Human | 肺泡灌洗液 | 在重症监护室的新冠病毒感染患者中,中性粒细胞高度激活 | EGAS00001004571 (EGA) |
| 人Human | 皮肤 | 新冠病毒感染引起皮疹机制 | GSE161225 |
| 人Human | 血液、鼻上皮、唾液 | CCL3等基因为严重新冠病毒感染鼻腔、口腔和血液中的生物标志物 | GSE183071 |
| 人Human | 上呼吸道、鼻咽、全血 | 发现与新冠病毒感染每种分子亚型相关的特定基因 | GSE156063; GSE163151 |
人Human | 外周血、全血、血浆和白细胞 | ARG1等7个基因生物标志物可区分SARS-CoV-2相关的ARI和其他类型的ARI | GSE161731; GSE171110 GSE152641; GSE157103 GSE172114; GSE161777 |
| 人Human | PBMC、全血、血浆和白细胞 | 生物标志物TGFBI、TTYH2和CD4参与免疫反应和炎症 | GSE152418; GSE166424; GSE157103 |
| 人Human | 全血 | 新冠病毒感染严重程度的转折点与S100A9等8个中性粒细胞特征基因有关 | E-MTAB-11240 (EMBL-EBI) |
| 人Human | 外周血 | 神经损伤相关的UCHL1表达增加与严重症状有关 | GSE152418 |
| 人Human | 外周血 | miR-146-146a-5p等3个非编码RNA是新冠病毒感染的潜在治疗靶点 | HRA000238 (NGDC) |
| 人Human | 外周血 | 鉴定了6个具有不同分子特征的新冠肺炎亚型 | GSE143507 |
| 人Human | 血沉棕黄层细胞、A549细胞 | cfDNA能够作为查看疾病严重程度的非侵入性生物标志物 | GSE147507; GSE154998 |
| 人Human | PBMC | 无症状患者中PBMC的免疫反应较弱,复阳患者的PBMC具有高炎症免疫反应 | GSE179627 |
| 人Human | PBMC | 循环单核淋巴细胞的变化可预测新冠病毒感染的严重程度 | GSE180594 |
| 人Human | 额叶皮质、脉络丛 | 新冠病毒感染患者脑和脉络丛细胞的功能失调 | GSE159812 |
表1 SARS-CoV-2感染转录组临床意义挖掘及对应数据集
Table 1 Clinical significances of SARS-CoV-2 transcriptome and associated datasets
样本来源 Sample origins | 组织/细胞 Tissues/cells | 发现 Finding | 数据集编号 Datasets' number |
|---|---|---|---|
人;雪貂 Human; ferrets | NHBE、A549、Calu3细胞,肺组织(人); 鼻腔冲洗液(雪貂) | 新冠病毒感染肺损伤相关机制 | GSE147507 |
人;雪貂 Human; ferrets | NHBE、A549、Calu3细胞(人);鼻腔冲洗 液(雪貂) | 应对新冠病毒感染时NHBE细胞上调MALAT1和NEAT1表达 | GSE147507 |
人;C57BL/6小鼠 Human; C57BL/6 mice | PBMC、肺泡灌洗液(人);淋巴结(小鼠) | 髓细胞、上皮细胞和T细胞之间的相互作用驱动组织损伤 | GSE149689; GSE155673; GSE145926; GSE149443 |
金黄仓鼠 Golden hamsters | 心脏 | 新冠病毒感染对心脏的损伤机制 | PRJNA884511 |
| 人Human | 肺、肝脏 | 新冠病毒感染肝损伤相关机制 | GSE150316 |
| 人Human | 肺泡灌洗液 | 严重新冠病毒感染患者的支气管肺泡灌洗液中富含促炎单核细胞衍生的巨噬细胞 | GSE145926 |
| 人Human | 肺泡灌洗液 | M1型巨噬细胞和重度新冠病毒感染患者之间的关系最高 | GSE145926 |
| 人Human | 肺泡灌洗液 | 在重症监护室的新冠病毒感染患者中,中性粒细胞高度激活 | EGAS00001004571 (EGA) |
| 人Human | 皮肤 | 新冠病毒感染引起皮疹机制 | GSE161225 |
| 人Human | 血液、鼻上皮、唾液 | CCL3等基因为严重新冠病毒感染鼻腔、口腔和血液中的生物标志物 | GSE183071 |
| 人Human | 上呼吸道、鼻咽、全血 | 发现与新冠病毒感染每种分子亚型相关的特定基因 | GSE156063; GSE163151 |
人Human | 外周血、全血、血浆和白细胞 | ARG1等7个基因生物标志物可区分SARS-CoV-2相关的ARI和其他类型的ARI | GSE161731; GSE171110 GSE152641; GSE157103 GSE172114; GSE161777 |
| 人Human | PBMC、全血、血浆和白细胞 | 生物标志物TGFBI、TTYH2和CD4参与免疫反应和炎症 | GSE152418; GSE166424; GSE157103 |
| 人Human | 全血 | 新冠病毒感染严重程度的转折点与S100A9等8个中性粒细胞特征基因有关 | E-MTAB-11240 (EMBL-EBI) |
| 人Human | 外周血 | 神经损伤相关的UCHL1表达增加与严重症状有关 | GSE152418 |
| 人Human | 外周血 | miR-146-146a-5p等3个非编码RNA是新冠病毒感染的潜在治疗靶点 | HRA000238 (NGDC) |
| 人Human | 外周血 | 鉴定了6个具有不同分子特征的新冠肺炎亚型 | GSE143507 |
| 人Human | 血沉棕黄层细胞、A549细胞 | cfDNA能够作为查看疾病严重程度的非侵入性生物标志物 | GSE147507; GSE154998 |
| 人Human | PBMC | 无症状患者中PBMC的免疫反应较弱,复阳患者的PBMC具有高炎症免疫反应 | GSE179627 |
| 人Human | PBMC | 循环单核淋巴细胞的变化可预测新冠病毒感染的严重程度 | GSE180594 |
| 人Human | 额叶皮质、脉络丛 | 新冠病毒感染患者脑和脉络丛细胞的功能失调 | GSE159812 |
物种 Species | 常用品种/品系 Breeds/ strains in common use | 感染滴度或 剂量 Infection titer or dose | 感染途径 Routes of infection | 用途 Applications | 优点 Advantages | 缺点 Disadvantages | GEO数据集数量 Number of GEO datasets |
|---|---|---|---|---|---|---|---|
小鼠 Mice | hACE2转基因 小鼠 | 1×105 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 方便,便宜 | 耗时 | 21 |
小鼠 Mice | K18-hACE2小鼠 | 1×102 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 方便,便宜 | 非致死模型 | 9 |
小鼠 Mice | hACE2-KI人源 化小鼠 | 4×105 PFU | 滴鼻 | 病毒感染抗体评价,疫苗评价 | 方便,便宜 | 非致死模型 | - |
仓鼠 Hamsters | 金黄地鼠 | 8×104 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 便宜,有消化系统症状 | 操作不如小鼠方便,病毒7 d清除,不致死 | 41 |
雪貂 Ferrets | - | 1×105 PFU | 滴鼻 | 致病机制研究,疫苗和药物评估 | 能通过直接接触和气溶胶感染 | 检测不如小鼠经济 | 5 |
猴 Monkeys | 恒河猴 | 1×106 TCID50 | 经气管感染 | 致病机制研究,疫苗和药物评估 | 与人类高度相似 | 价格昂贵 | 25 |
猴 Monkeys | 食蟹猴 | 1×106 PFU | 气管、鼻腔、黏膜 联合感染 | 评价新冠药物与疫苗 | 与人类高度相似 | 价格昂贵 | 8 |
水貂 Minks | - | 1×106 TCID50 | 经气管感染 | 临床病理机制阐述,药物和疫苗评价,生物标志物筛选 | 与人类症状较为 相似 | 应用少,相关试剂较少 | - |
树鼩 Tree shrews | - | 1×107 TCID50 | 口、气管、眼联合 感染 | 药物筛选和疫苗评估 | 便宜,与灵长类 相似 | 没有纯遗传背景的品系,个体差异较大 | - |
表2 不同物种的新冠病毒感染性差异
Table 2 Infectious differences of SARS-CoV-2 in different species
物种 Species | 常用品种/品系 Breeds/ strains in common use | 感染滴度或 剂量 Infection titer or dose | 感染途径 Routes of infection | 用途 Applications | 优点 Advantages | 缺点 Disadvantages | GEO数据集数量 Number of GEO datasets |
|---|---|---|---|---|---|---|---|
小鼠 Mice | hACE2转基因 小鼠 | 1×105 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 方便,便宜 | 耗时 | 21 |
小鼠 Mice | K18-hACE2小鼠 | 1×102 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 方便,便宜 | 非致死模型 | 9 |
小鼠 Mice | hACE2-KI人源 化小鼠 | 4×105 PFU | 滴鼻 | 病毒感染抗体评价,疫苗评价 | 方便,便宜 | 非致死模型 | - |
仓鼠 Hamsters | 金黄地鼠 | 8×104 TCID50 | 滴鼻 | 致病机制研究,疫苗和药物评估 | 便宜,有消化系统症状 | 操作不如小鼠方便,病毒7 d清除,不致死 | 41 |
雪貂 Ferrets | - | 1×105 PFU | 滴鼻 | 致病机制研究,疫苗和药物评估 | 能通过直接接触和气溶胶感染 | 检测不如小鼠经济 | 5 |
猴 Monkeys | 恒河猴 | 1×106 TCID50 | 经气管感染 | 致病机制研究,疫苗和药物评估 | 与人类高度相似 | 价格昂贵 | 25 |
猴 Monkeys | 食蟹猴 | 1×106 PFU | 气管、鼻腔、黏膜 联合感染 | 评价新冠药物与疫苗 | 与人类高度相似 | 价格昂贵 | 8 |
水貂 Minks | - | 1×106 TCID50 | 经气管感染 | 临床病理机制阐述,药物和疫苗评价,生物标志物筛选 | 与人类症状较为 相似 | 应用少,相关试剂较少 | - |
树鼩 Tree shrews | - | 1×107 TCID50 | 口、气管、眼联合 感染 | 药物筛选和疫苗评估 | 便宜,与灵长类 相似 | 没有纯遗传背景的品系,个体差异较大 | - |
物种 Species | 病毒复制部位 Virus replication site | 病理变化 Pathological change | 免疫应答 Immune response | 参考文献 References |
|---|---|---|---|---|
人类 Human | 主要在呼吸道和肠道 | 肺炎,急性呼吸窘迫综合征 | 产生针对S和N蛋白以及RBD的IgG抗体 | [ |
小鼠(hACE2转基因小鼠) Mice( hACE2 transgenic mice ) | 肺 | 间质性肺炎 | 产生抗S蛋白的特异性抗体 | [ |
仓鼠 Hamsters | 肺、鼻甲、气管、肠道 | 肺炎,弥漫性肺泡损伤 | 中和抗体反应,IFN-R、IL-4、IL-6、TNF-a、 IL-12p40上调中和抗体反应 | [ |
雪貂 Ferrets | 鼻甲、气管、肺、肠道 | 毛细支气管炎 | - | [ |
水貂 Minks | 鼻腔、直肠、耳 | 间质性肺炎,血管周炎,细支气 管周炎,鼻黏膜损伤 | - | [ |
恒河猴 Rhesus monkey | 肺、气管、支气管 | 间质性肺炎 | 白细胞和中性粒细胞减少,淋巴细胞增加 | [ |
表3 不同物种的新冠病毒致病性差异
Table 3 Pathogenic differences of SARS-CoV-2 infection in different species
物种 Species | 病毒复制部位 Virus replication site | 病理变化 Pathological change | 免疫应答 Immune response | 参考文献 References |
|---|---|---|---|---|
人类 Human | 主要在呼吸道和肠道 | 肺炎,急性呼吸窘迫综合征 | 产生针对S和N蛋白以及RBD的IgG抗体 | [ |
小鼠(hACE2转基因小鼠) Mice( hACE2 transgenic mice ) | 肺 | 间质性肺炎 | 产生抗S蛋白的特异性抗体 | [ |
仓鼠 Hamsters | 肺、鼻甲、气管、肠道 | 肺炎,弥漫性肺泡损伤 | 中和抗体反应,IFN-R、IL-4、IL-6、TNF-a、 IL-12p40上调中和抗体反应 | [ |
雪貂 Ferrets | 鼻甲、气管、肺、肠道 | 毛细支气管炎 | - | [ |
水貂 Minks | 鼻腔、直肠、耳 | 间质性肺炎,血管周炎,细支气 管周炎,鼻黏膜损伤 | - | [ |
恒河猴 Rhesus monkey | 肺、气管、支气管 | 间质性肺炎 | 白细胞和中性粒细胞减少,淋巴细胞增加 | [ |
类别 Category | 内容 Content | 数量 Number |
|---|---|---|
物种 Species | 人、雪貂 | 2 |
病毒亚型 Virus subtypes | 野生型、USA-WA1型 | 2 |
细胞/组织类型 Tissue/ cell types | 肺、气管、鼻腔,肺癌Calu3、A549、A549_oeACE2、H1299细胞,结直肠癌Caco2细胞,正常人类支气管上皮细胞 | 9 |
时间点 Time point | 4 h、8 h、12 h、24 h、36 h、72 h、7 d、14 d | 8 |
感染剂量 Infection dose | 0 MOI、0.2 MOI、0.3 MOI、2 MOI、5×104 PFU | 5 |
数据集编号 Dataset number | GSE147507,GSE148729 | 2 |
表4 有关冠状病毒感染动物模型的COVID-CED数据整理
Table 4 Data collation of COVID-CED related to SARS-CoV-2 infectious animal model
类别 Category | 内容 Content | 数量 Number |
|---|---|---|
物种 Species | 人、雪貂 | 2 |
病毒亚型 Virus subtypes | 野生型、USA-WA1型 | 2 |
细胞/组织类型 Tissue/ cell types | 肺、气管、鼻腔,肺癌Calu3、A549、A549_oeACE2、H1299细胞,结直肠癌Caco2细胞,正常人类支气管上皮细胞 | 9 |
时间点 Time point | 4 h、8 h、12 h、24 h、36 h、72 h、7 d、14 d | 8 |
感染剂量 Infection dose | 0 MOI、0.2 MOI、0.3 MOI、2 MOI、5×104 PFU | 5 |
数据集编号 Dataset number | GSE147507,GSE148729 | 2 |
| 1 | LOWE R, SHIRLEY N, BLEACKLEY M, et al. Transcriptomics technologies[J]. PLoS Comput Biol, 2017, 13(5): e1005457. DOI: 10.1371/journal.pcbi.1005457 . |
| 2 | 国家卫生健康委. «关于对新型冠状病毒感染实施"乙类乙管"的总体方案»解读问答[A/OL]. [2023-10-01]. . |
| National Health Commission of the People's Republic of China. Questions and answers on "overall plan of implementing 'class B management' for novel coronavirus infection"[J]. [A/OL]. [2023-10-01]. . | |
| 3 | JHA P K, VIJAY A, HALU A, et al. Gene expression profiling reveals the shared and distinct transcriptional signatures in human lung epithelial cells infected with SARS-CoV-2, MERS-CoV, or SARS-CoV: potential implications in cardiovascular complications of COVID-19[J]. Front Cardiovasc Med, 2021, 7:623012. DOI: 10.3389/fcvm.2020.623012 . |
| 4 | CROSS A R, DE ANDREA C E, VILLALBA-ESPARZA M, et al. Spatial transcriptomic characterization of COVID-19 pneumonitis identifies immune circuits related to tissue injury[J]. JCI Insight, 2023, 8(2): e157837. DOI: 10.1172/jci.insight.157837 . |
| 5 | HAMMOUDEH S M, HAMMOUDEH A M, BHAMIDIMARRI P M, et al. Insight into molecular mechanisms underlying hepatic dysfunction in severe COVID-19 patients using systems biology[J]. World J Gastroenterol, 2021, 27(21):2850-2870. DOI: 10.3748/wjg.v27.i21.2850 . |
| 6 | MITAMURA Y, SCHULZ D, ORO S, et al. Cutaneous and systemic hyperinflammation drives maculopapular drug exanthema in severely ill COVID-19 patients[J]. Allergy, 2022, 77(2):595-608. DOI: 10.1111/all.14983 . |
| 7 | HIRSCH J S, NG J H, ROSS D W, et al. Acute kidney injury in patients hospitalized with COVID-19[J]. Kidney Int, 2020, 98(1):209-218. DOI: 10.1016/j.kint.2020.05.006 . |
| 8 | ALEXANDER M P, MANGALAPARTHI K K, MADUGUNDU A K, et al. Acute kidney injury in severe COVID-19 has similarities to sepsis-associated kidney injury: a multi-omics study[J]. Mayo Clin Proc, 2021, 96(10):2561-2575. DOI: 10.1016/j.mayocp.2021.07.001 . |
| 9 | LONG B, BRADY W J, KOYFMAN A, et al. Cardiovascular complications in COVID-19[J]. Am J Emerg Med, 2020, 38(7):1504-1507. DOI: 10.1016/j.ajem.2020.04.048 . |
| 10 | WISCOVITCH-RUSSO R, IBÁÑEZ-PRADA E D, SERRANO-MAYORGA C C, et al. Major adverse cardiovascular events are associated with necroptosis during severe COVID-19[J]. Crit Care, 2023, 27(1):155. DOI: 10.1186/s13054-023-04423-8 . |
| 11 | CHEN X P, WU T, LI L G, et al. Transcriptional start site coverage analysis in plasma cell-free DNA reveals disease severity and tissue specificity of COVID-19 patients[J]. Front Genet, 2021, 12:663098. DOI: 10.3389/fgene.2021.663098 . |
| 12 | GÓMEZ-CARBALLA A, RIVERO-CALLE I, PARDO-SECO J, et al. A multi-tissue study of immune gene expression profiling highlights the key role of the nasal epithelium in COVID-19 severity[J]. Environ Res, 2022, 210:112890. DOI: 10.1016/j.envres.2022.112890 . |
| 13 | KRISHNAMOORTHY P, RAJ A S, KUMAR H. Machine learning-driven blood transcriptome-based discovery of SARS-CoV-2 specific severity biomarkers[J]. J Med Virol, 2023, 95(2): e28488. DOI: 10.1002/jmv.28488 . |
| 14 | CLANCY J, HOFFMANN C S, PICKETT B E. Transcriptomics secondary analysis of severe human infection with SARS-CoV-2 identifies gene expression changes and predicts three transcriptional biomarkers in leukocytes[J]. Comput Struct Biotechnol J, 2023, 21:1403-1413. DOI: 10.1016/j.csbj.2023.02.003 . |
| 15 | LIU Y, WU Y K, LIU B, et al. Biomarkers and immune repertoire metrics identified by peripheral blood transcriptomic sequencing reveal the pathogenesis of COVID-19[J]. Front Immunol, 2021, 12:677025. DOI: 10.3389/fimmu.2021.677025 . |
| 16 | ASCHENBRENNER A C, MOUKTAROUDI M, KRÄMER B, et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients[J]. Genome Med, 2021, 13(1):7. DOI: 10.1186/s13073-020-00823-5 . |
| 17 | ZHANG J Q, LIN D Z, LI K, et al. Transcriptome analysis of peripheral blood mononuclear cells reveals distinct immune response in asymptomatic and re-detectable positive COVID-19 patients[J]. Front Immunol, 2021, 12:716075. DOI: 10.3389/fimmu.2021.716075 . |
| 18 | MUKHERJEE S, BANERJEE B, KARASIK D, et al. mRNA-lncRNA co-expression network analysis reveals the role of lncRNAs in immune dysfunction during severe SARS-CoV-2 infection[J]. Viruses, 2021, 13(3):402. DOI: 10.3390/v13030402 . |
| 19 | KARAMI H, DERAKHSHANI A, GHASEMIGOL M, et al. Weighted gene co-expression network analysis combined with machine learning validation to identify key modules and hub genes associated with SARS-CoV-2 infection[J]. J Clin Med, 2021, 10(16):3567. DOI: 10.3390/jcm10163567 . |
| 20 | HU R W, LIU C, YAN Y Y, et al. Identification of hub genes and molecular subtypes in COVID-19 based on WGCNA[J]. Eur Rev Med Pharmacol Sci, 2021, 25(20):6411-6424. DOI: 10.26355/eurrev_202110_27015 . |
| 21 | PANIRI A, AKHAVAN-NIAKI H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: role of lncRNAs in cytokine storm modulation[J]. Life Sci, 2020, 257:118114. DOI: 10.1016/j.lfs.2020.118114 . |
| 22 | VISHNUBALAJI R, SHAATH H, ALAJEZ N M. Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS-CoV-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response[J]. Genes, 2020, 11(7):760. DOI: 10.3390/genes11070760 . |
| 23 | TANG H, GAO Y H, LI Z H, et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19[J]. Clin Transl Med, 2020, 10(6): e200. DOI: 10.1002/ctm2.200 . |
| 24 | LIAO M F, LIU Y, YUAN J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19[J]. Nat Med, 2020, 26(6):842-844. DOI: 10.1038/s41591-020-0901-9 . |
| 25 | ZHANG Z L, CUI F F, CAO C, et al. Single-cell RNA analysis reveals the potential risk of organ-specific cell types vulnerable to SARS-CoV-2 infections[J]. Comput Biol Med, 2021, 140:105092. DOI: 10.1016/j.compbiomed.2021.105092 . |
| 26 | FILBIN M R, MEHTA A, SCHNEIDER A M, et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions[J]. Cell Rep Med, 2021, 2(5):100287. DOI: 10.1016/j.xcrm.2021.100287 . |
| 27 | FUZO C A, FRAGA-SILVA T F C, MARUYAMA S R, et al. The turning point of COVID-19 severity is associated with a unique circulating neutrophil gene signature[J]. Immunology, 2023, 169(3):323-343. DOI: 10.1111/imm.13631 . |
| 28 | SCHIMKE L F, MARQUES A H C, BAIOCCHI G C, et al. Severe COVID-19 shares a common neutrophil activation signature with other acute inflammatory states[J]. Cells, 2022, 11(5):847. DOI: 10.3390/cells11050847 . |
| 29 | UTRERO-RICO A, GONZÁLEZ-CUADRADO C, CHIVITE-LACABA M, et al. Alterations in circulating monocytes predict COVID-19 severity and include chromatin modifications still detectable six months after recovery[J]. Biomedicines, 2021, 9(9):1253. DOI: 10.3390/biomedicines 9091253 . |
| 30 | YANG A C, KERN F, LOSADA P M, et al. Publisher Correction: Dysregulation of brain and choroid plexus cell types in severe COVID-19[J]. Nature, 2021, 598(7882): E4. DOI: 10.1038/s41586-021-04080-3 . |
| 31 | 吴玥, 薛婧, 魏强, 等. 国家动物模型资源共享信息平台的建立[J]. 中国实验动物学报, 2022, 30(8): 1080-1086. DOI: 10.3969/j.issn.1005-4847.2022.08.009 . |
| WU Y, XUE J, WEI Q, et al. Establishment of national infrastructure for an animal model resource platform[J]. Acta Lab Animalis Sci Sin, 2022, 30(8): 1080-1086. DOI: 10.3969/j.issn.1005-4847.2022.08.009 . | |
| 32 | ZHENG J X, DENG Y Y, ZHAO Z Y, et al. Characterization of SARS-CoV-2-specific humoral immunity and its potential applications and therapeutic prospects[J]. Cell Mol Immunol, 2022, 19(2):150-157. DOI: 10.1038/s41423-021-00774-w . |
| 33 | BAO L L, DENG W, HUANG B Y, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice[J]. Nature, 2020, 583(7818):830-833. DOI: 10.1038/s41586-020-2312-y . |
| 34 | CHAN J F W, ZHANG A J, YUAN S F, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian Hamster model: implications for disease pathogenesis and transmissibility[J]. Clin Infect Dis, 2020, 71(9):2428-2446. DOI: 10.1093/cid/ciaa325 . |
| 35 | KIM Y I, KIM S G, KIM S M, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets[J]. Cell Host Microbe, 2020, 27(5):704-709.e2. DOI: 10.1016/j.chom.2020.03.023 . |
| 36 | SHUAI L, ZHONG G X, YUAN Q, et al. Replication, pathogenicity, and transmission of SARS-CoV-2 in minks[J]. Natl Sci Rev, 2021, 8(3): nwaa291. DOI: 10.1093/nsr/nwaa291 . |
| 37 | SHAN C, YAO Y F, YANG X L, et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques[J]. Cell Res, 2020, 30(8):670-677. DOI: 10.1038/s41422-020-0364-z . |
| 38 | 吴玥, 向志光, 高苒, 等. 冠状病毒感染动物模型比较转录组学数据库的建立[J]. 中国实验动物学报, 2022, 30(1):92-99. DOI: 10.3969/j.issn.1005-4847.2022.01.012 . |
| WU Y, XIANG Z G, GAO R, et al. Establishment of a comparative transcriptomics database of coronavirus infected animal models[J]. Acta Lab Anim Sci Sinica, 2022, 30(1):92-99. DOI: 10.3969/j.issn.1005-4847.2022.01.012 . | |
| 39 | REN X W, WEN W, FAN X Y, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas[J]. Cell, 2021, 184(7):1895-1913.e19. DOI: 10.1016/j.cell.2021.01.053 . |
| 40 | MECKIFF B J, RAMÍREZ-SUÁSTEGUI C, FAJARDO V, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19[J]. Cell, 2020, 183(5):1340-1353.e16. DOI: 10.1016/j.cell.2020.10.001 . |
| 41 | NAGAFUCHI Y, YANAOKA H, FUJIO K. Lessons from transcriptome analysis of autoimmune diseases[J]. Front Immunol, 2022, 13:857269. DOI: 10.3389/fimmu.2022.857269 . |
| 42 | ROBINSON E L, BAKER A H, BRITTAN M, et al. Dissecting the transcriptome in cardiovascular disease[J]. Cardiovasc Res, 2022, 118(4):1004-1019. DOI: 10.1093/cvr/cvab117 . |
| 43 | PARK H W, WEISS S T. Understanding the molecular mechanisms of asthma through transcriptomics[J]. Allergy Asthma Immunol Res, 2020, 12(3):399-411. DOI: 10.4168/aair.2020.12.3.399 . |
| [1] | 张渊, 李晗, 张成芳. 奥氮平诱导体重增加小鼠的全脑转录组学分析[J]. 实验动物与比较医学, 2023, 43(3): 262-270. |
| [2] | 杨凌, 庄迪, 金立伦. 转录组测序筛选大鼠滑膜炎差异表达基因及秦皮素治疗靶点的体外验证[J]. 实验动物与比较医学, 2023, 43(1): 11-20. |
| [3] | 温金银, 陈焰, 李会萍. 实验动物电商平台数据分析与对策建议[J]. 实验动物与比较医学, 2021, 41(4): 363-368. |
| [4] | 郑杰, 张国忠, 胡艳梅, 伍学一, 杨宗富, 字向东. 基于RNA-Seq技术的犏牛囊胚冷冻前后单核苷酸多态性和可变剪切分析[J]. 实验动物与比较医学, 2020, 40(4): 279-. |
| [5] | 王丰, 郝飞, 还红华, 张元鹏, 张波, 邵国青. 日本大耳白兔和新西兰兔体温周期变动规律的研究[J]. 实验动物与比较医学, 2015, 35(3): 222-226. |
| [6] | 詹瑧,侯亮. 表皮生长因子影响小鼠舌苔形成的信号传导分子机制初探[J]. , 2003, 23(1): 7-9. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||