
Laboratory Animal and Comparative Medicine ›› 2025, Vol. 45 ›› Issue (2): 147-157.DOI: 10.12300/j.issn.1674-5817.2024.119
• Animal Models of Human Diseases • Previous Articles Next Articles
PAN Qianjia1, GE Junyi1, HU Nan2, HUA Fei3, GU Min1( )
)
Received:2024-08-21
Revised:2024-12-24
Online:2025-04-25
Published:2025-04-25
Contact:
GU Min
CLC Number:
PAN Qianjia,GE Junyi,HU Nan,et al. Differential Analysis of Oral Microbiota in db/db Mouse Model of Type 2 Diabetes Utilizing 16S rRNA Sequencing[J]. Laboratory Animal and Comparative Medicine, 2025, 45(2): 147-157. DOI: 10.12300/j.issn.1674-5817.2024.119.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2024.119
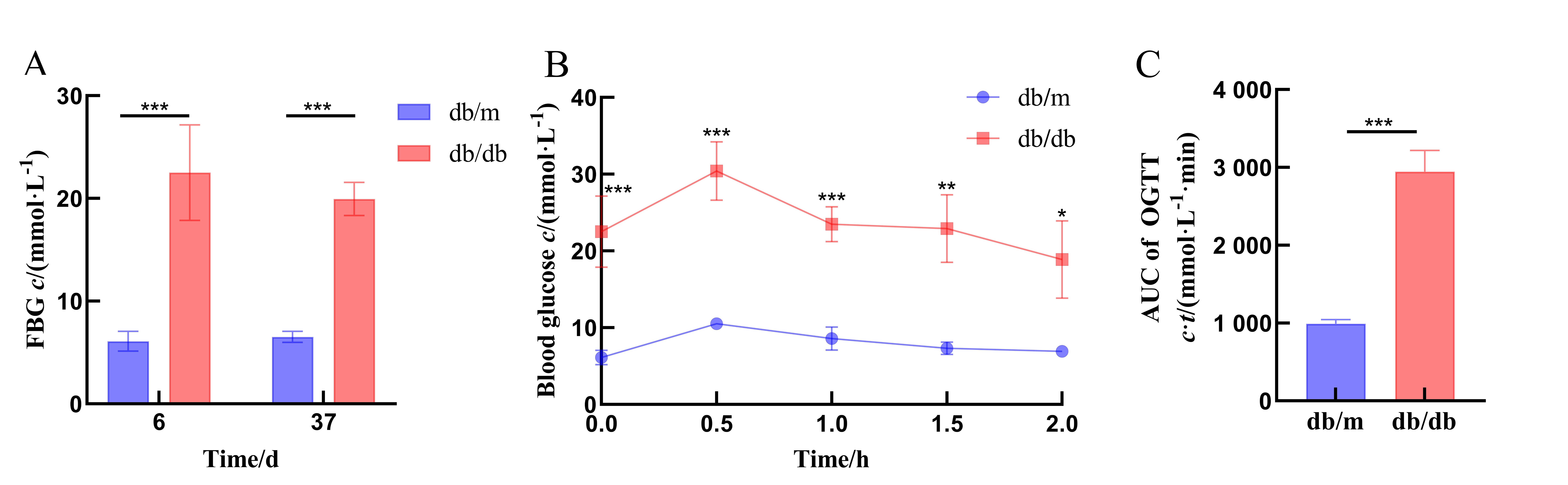
Figure 1 Comparison of blood glucose indices in db/db mice and db/m miceNote:A, The concentration of fasting blood glucose (FBG) levels after a 12-hour fasting period; B, Analysis of oral glucose tolerance test (OGTT) from 0-2 hours; C, Area under the curve (AUC) of the OGTT. Db/db mice served as the diabetic group, while db/m mice were the normal controls, n=8 per group. *P<0.05, **P<0.01, ***P<0.001.
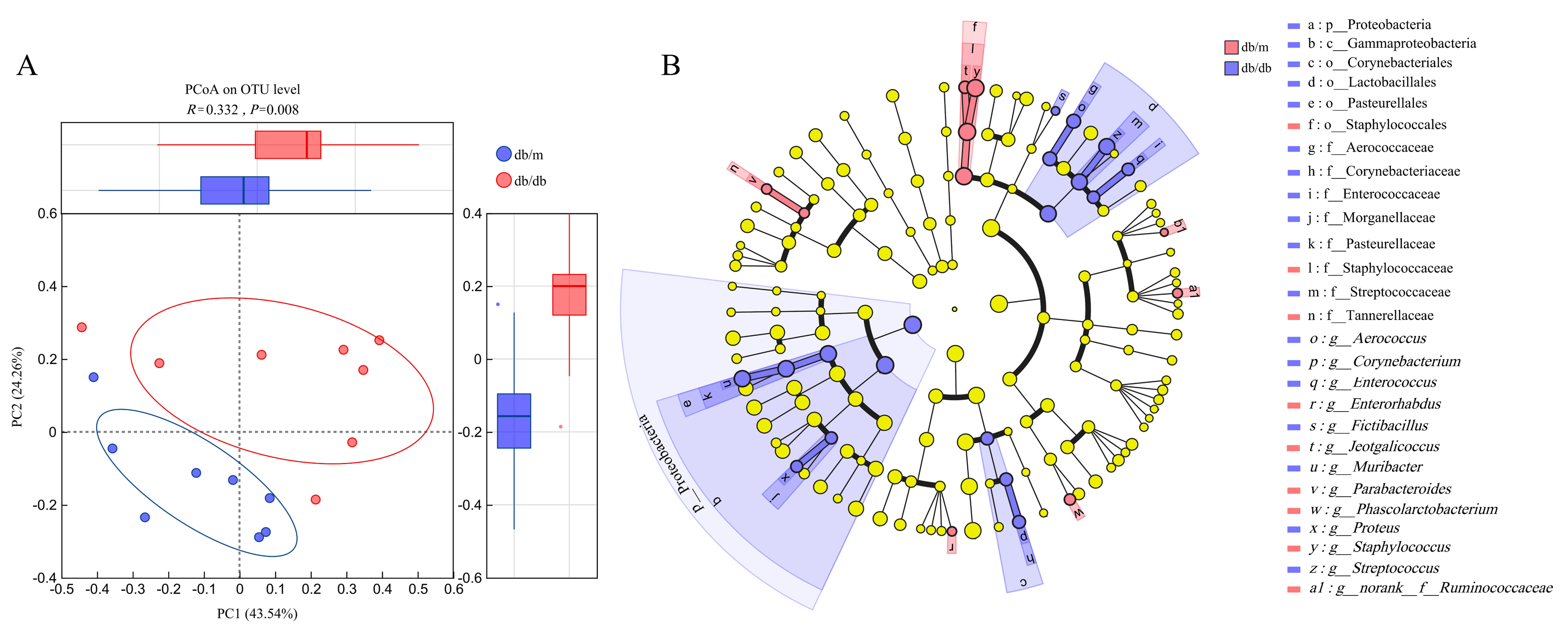
Figure 2 PCoA and LEfSe analysis of oral microbiota composition in db/db mice and db/m miceNote:A, Principal coordinate analysis (PCoA) between db/db mice and db/m mice; B, LEfSe-derived cladogram showing taxonomic differences from phylum to genus level (p for phylum; c for class; o for order; f for family; g for genus). Db/db mice served as the diabetic model, while db/m mice were the normal controls, n=8 per group.
指标 Index | db/m | db/db | P value |
|---|---|---|---|
观察物种指数 Sobs | 10.25±8.43 | 77.25±29.33 | 0.049* |
香农指数# Shannon# | 2.40 (2.25~2.48) | 2.32 (2.23~2.51) | 0.529 |
辛普森指数 Simpson | 0.18±0.04* | 0.16±0.04 | 0.499 |
基于丰度的覆盖估计值 ACE | 105.56±8.34 | 79.49±30.06 | 0.045* |
Chao1丰富度估计量 Chao1 | 105.67±9.78 | 79.25±30.42 | 0.046* |
测序覆盖度 Coverage | 0.999 9 | 1.000 0 | 0.458 |
Table 1 Alpha (α) diversity analysis of oral microbiota in db/db mice and db/m mice
指标 Index | db/m | db/db | P value |
|---|---|---|---|
观察物种指数 Sobs | 10.25±8.43 | 77.25±29.33 | 0.049* |
香农指数# Shannon# | 2.40 (2.25~2.48) | 2.32 (2.23~2.51) | 0.529 |
辛普森指数 Simpson | 0.18±0.04* | 0.16±0.04 | 0.499 |
基于丰度的覆盖估计值 ACE | 105.56±8.34 | 79.49±30.06 | 0.045* |
Chao1丰富度估计量 Chao1 | 105.67±9.78 | 79.25±30.42 | 0.046* |
测序覆盖度 Coverage | 0.999 9 | 1.000 0 | 0.458 |
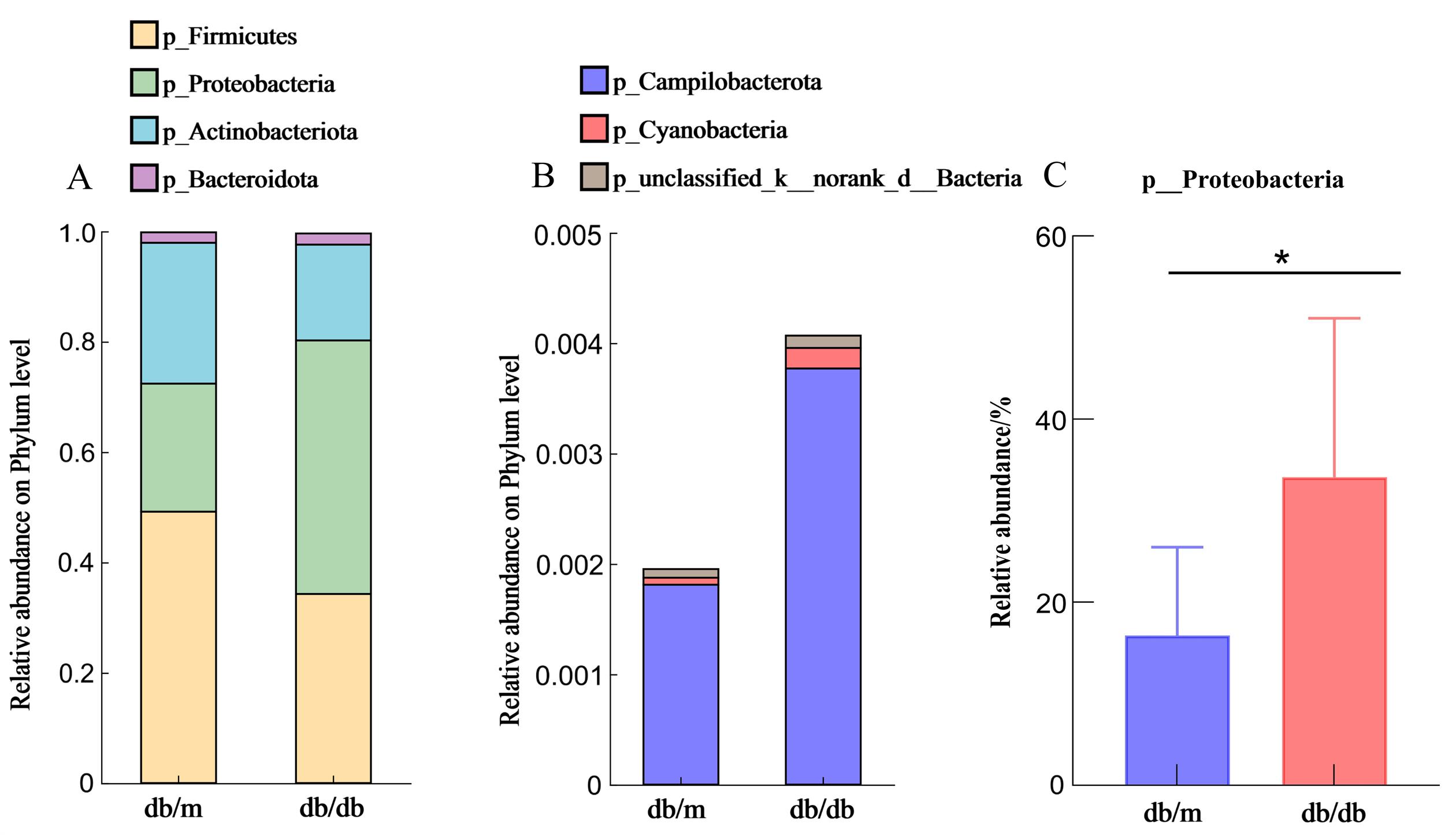
Figure 3 Comparison of oral microbiota between db/db mice and db/m mice at the phylum levelNote:A-B, Phylum-level community composition of oral microbiota in db/db and db/m groups; C, Relative abundance of p_Proteobacteria. db/db mice served as the diabetic model, while db/m mice were the normal controls, n=8 per group. *P<0.05.
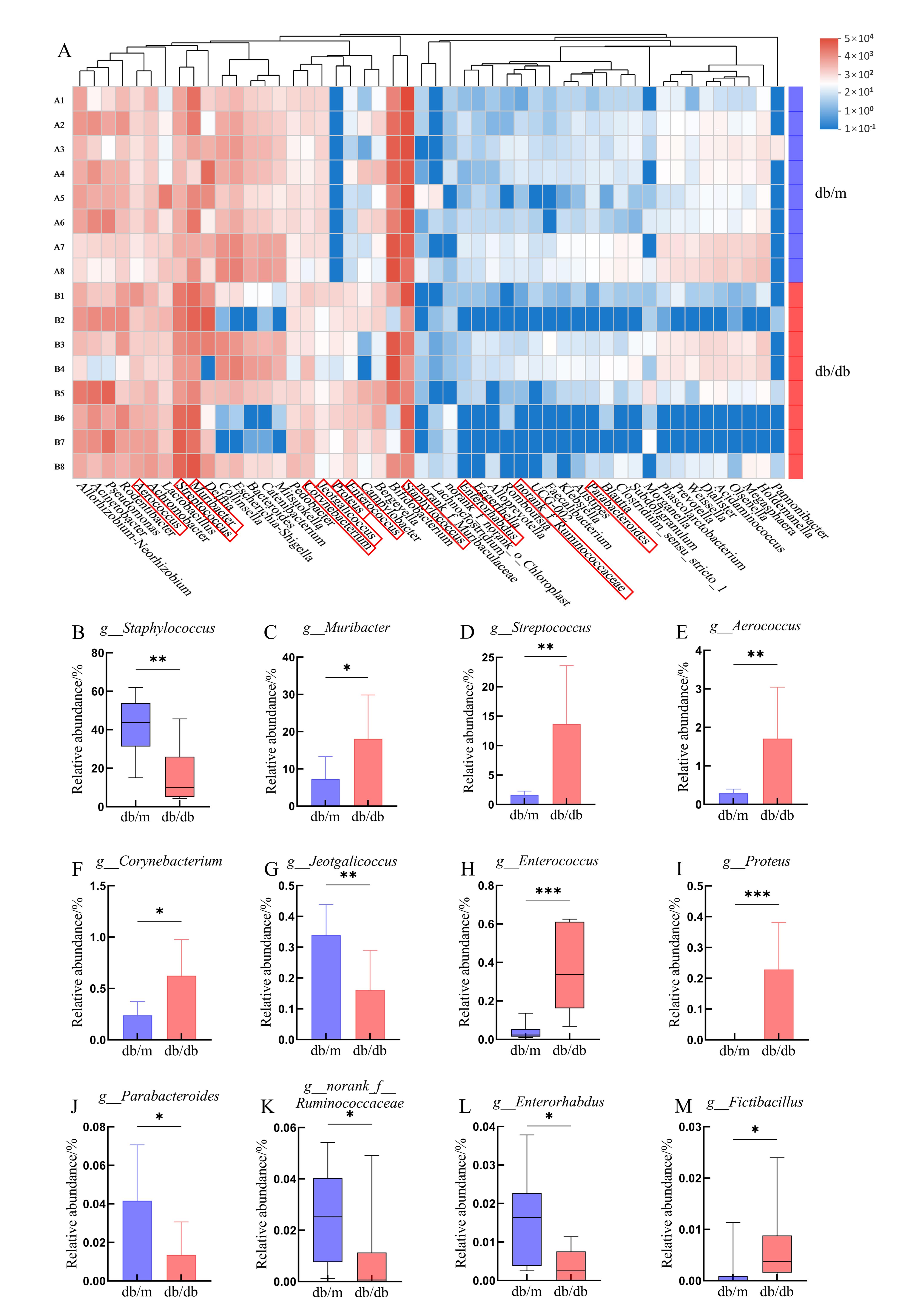
Figure 4 Differences in oral microbiota composition between db/db mice and db/m mice at the genus levelNote: A, Heatmap showing the relative abundance of the top 50 dominant species in the saliva microbiota (db/db vs. db/m); B-M, Relative abundance of the following genera: g_Staphylococcus, g_Muribacter, g_Streptococcus, g_Aerococcus, g_Corynebacterium, g_Jeotgalicoccus, g_Enterococcus, g_Proteus, g_Parabacteroides, g_Ruminococcaceae, g_Enterorhabdus, and g_Fictibacillus. Normally distributed data are presented as bar plots, and non-normally distributed data as box plots. Db/db mice served as the diabetic model, while db/m mice were the normal controls, n=8 per group. *P < 0.05, **P < 0.01, ***P < 0.001.
| 1 | RUSSO M P, GRANDE-RATTI M F, BURGOS M A, et al. Prevalence of diabetes, epidemiological characteristics and vascular complications[J]. Arch Cardiol Mex, 2023, 93(1):30-36. DOI:10.24875/ACM.21000410 . |
| 2 | RUZE R, LIU T T, ZOU X, et al. Obesity and type 2 diabetes mellitus: connections in epidemiology, pathogenesis, and treatments[J]. Front Endocrinol, 2023, 14:1161521. DOI:10.3389/fendo.2023.1161521 . |
| 3 | ZHENG Y, LEY S H, HU F B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications[J]. Nat Rev Endocrinol, 2018, 14(2):88-98. DOI:10.1038/nrendo.2017.151 . |
| 4 | ZHANG Y, SONG M Y, CAO Y, et al. Incident early- and later-onset type 2 diabetes and risk of early- and later-onset cancer: prospective cohort study[J]. Diabetes Care, 2023, 46(1):120-129. DOI:10.2337/dc22-1575 . |
| 5 | MARTAGON A J, ZUBIRÁN R, GONZÁLEZ-ARELLANES R, et al. HDL abnormalities in type 2 diabetes: Clinical implications[J]. Atherosclerosis, 2024, 394:117213. DOI:10.1016/j.atherosclerosis.2023.117213 . |
| 6 | WANG D Y, ZHANG G S, YU Y N, et al. Imaging of sarcopenia in type 2 diabetes mellitus[J]. Clin Interv Aging, 2024, 19:141-151. DOI:10.2147/CIA.S443572 . |
| 7 | DENNIS J M, SHIELDS B M, HENLEY W E, et al. Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data[J]. Lancet Diabetes Endocrinol, 2019, 7(6):442-451. DOI:10.1016/S2213-8587(19)30087-7 . |
| 8 | EDDOUKS M, CHATTOPADHYAY D, ZEGGWAGH N A. Animal models as tools to investigate antidiabetic and anti-inflammatory plants[J]. Evid Based Complement Alternat Med, 2012, 2012:142087. DOI:10.1155/2012/142087 . |
| 9 | 唐艺丹, 王鲜忠, 张姣姣. Ⅱ型糖尿病动物模型构建的研究进展[J]. 中国实验动物学报, 2020, 28(6):870-876. DOI: 10.3969/j.issn.1005-4847.2020.06.020 . |
| TANG Y D, WANG X Z, ZHANG J J. Research progress in the construction of type Ⅱ diabetes animal models[J]. Acta Lab Animalis Sci Sin, 2020, 28(6):870-876. DOI: 10.3969/j.issn.1005-4847.2020.06.020 . | |
| 10 | 崔淼, 朱春江, 刘向荣. 糖尿病动物模型构建的研究进展[J]. 中国实验诊断学, 2023, 27(2):227-230. DOI:1007-4287(2023)02-0227-04 . |
| CUI M, ZHU C J, LIU X R. Research progress on the construction of animal model of diabetes mellitus[J]. Chin J Lab Diagn, 2023, 27(2):227-230. DOI:1007-4287(2023)02-0227-04 . | |
| 11 | WONG S K, CHIN K Y, SUHAIMI F H, et al. Animal models of metabolic syndrome: a review[J]. Nutr Metab, 2016, 13:65. DOI:10.1186/s12986-016-0123-9 . |
| 12 | 王松月, 依香叫, 李金诚, 等. 自发性2型糖尿病模型db/db小鼠生物学特性研究[J]. 中国中医基础医学杂志, 2019, 25(7):909-912. DOI: 10.19945/j.cnki.issn.1006-3250.2019.07.019 . |
| WANG S Y, YI X J, LI J C, et al. Biological characteristics of spontaneous type 2 diabetes mellitus model db/db mice[J]. Chin J Basic Med Tradit Chin Med, 2019, 25(7):909-912. DOI: 10.19945/j.cnki.issn.1006-3250.2019.07.019 . | |
| 13 | ZHANG Y H, WANG X, LI H X, et al. Human oral microbiota and its modulation for oral health[J]. Biomed Pharmacother, 2018, 99:883-893. DOI:10.1016/j.biopha.2018.01.146 . |
| 14 | 朱双, 汤帅, 丁刚. 口腔菌群与口腔疾病及全身性疾病关系的研究进展[J]. 中国医药导报, 2023, 20(18):35-38. DOI: 10.20047/j.issn1673-7210.2023.18.07 . |
| ZHU S, TANG S, DING G. Research progress on the relationship between oral flora and oral diseases and systemic diseases[J]. China Med Her, 2023, 20(18):35-38. DOI: 10.20047/j.issn1673-7210.2023.18.07 . | |
| 15 | MAIER T. Oral microbiome in health and disease: maintaining a healthy, balanced ecosystem and reversing dysbiosis[J]. Microorganisms, 2023, 11(6):1453. DOI:10.3390/micro-organisms11061453 . |
| 16 | LU Y B, LI Z Y, PENG X. Regulatory effects of oral microbe on intestinal microbiota and the illness[J]. Front Cell Infect Microbiol, 2023, 13:1093967. DOI:10.3389/fcimb.2023.1093967 . |
| 17 | TANG B Y, YAN C X, SHEN X, et al. The bidirectional biological interplay between microbiome and viruses in periodontitis and type-2 diabetes mellitus[J]. Front Immunol, 2022, 13:885029. DOI:10.3389/fimmu.2022.885029 . |
| 18 | STÖHR J, BARBARESKO J, NEUENSCHWANDER M, et al. Bidirectional association between periodontal disease and diabetes mellitus: a systematic review and meta-analysis of cohort studies[J]. Sci Rep, 2021, 11(1):13686. DOI:10.1038/s41598-021-93062-6 . |
| 19 | BARUTTA F, BELLINI S, DURAZZO M, et al. Novel insight into the mechanisms of the bidirectional relationship between diabetes and periodontitis[J]. Biomedicines, 2022, 10(1):178. DOI:10.3390/biomedicines10010178 . |
| 20 | SHINJO T, NISHIMURA F. The bidirectional association between diabetes and periodontitis, from basic to clinical[J]. Jpn Dent Sci Rev, 2024, 60:15-21. DOI:10.1016/j.jdsr.2023. 12.002 . |
| 21 | 杨忠良, 王健辉, 刘琳, 等. 实验性牙周炎对小鼠焦虑样行为及肠道菌群的影响[J]. 中华老年口腔医学杂志, 2023, 21(5):257-263. DOI: 10.19749/j.cn.cjgd.1672-2973.2023.05.001 . |
| YANG Z L, WANG J H, LIU L, et al. Effects of experimental periodontitis on anxiety-like behavior and gut microbiota in mice[J]. Chin J Geriatr Dent, 2023, 21(5):257-263. DOI: 10.19749/j.cn.cjgd.1672-2973.2023.05.001 . | |
| 22 | 刘静, 陆洁, 裴天仙, 等. 基因缺陷型2型糖尿病模型动物db/db小鼠早期生物学特性研究[J]. 药物评价研究, 2013, 36(1):8-12.DOI: 10.7501/i.issn.1674-6376.2013.01.003 . |
| LIU J, LU J, PEI T X, et al. Research on early biological characteristics of gene defect type 2 diabetes model db/db mice[J]. Drug Eval Res, 2013, 36(1):8-12. DOI: 10.7501/i.issn.1674-6376.2013.01.003 . | |
| 23 | 侯黎光, 叶宇, 苟惠清, 等. A20通过抑制自噬缓解实验性牙周炎牙槽骨吸收[J]. 南京医科大学学报(自然科学版), 2022, 42(4):476-483, 587. DOI: 10.7655/NYDXBNS20220403 . |
| HOU L G, YE Y, GOU H Q, et al. A20 alleviates alveolar bone loss in experimental periodontitis by inhibiting autophagy[J]. J Nanjing Med Univ (Nat Sci), 2022, 42(4):476-483, 587. DOI: 10.7655/NYDXBNS20220403 . | |
| 24 | KIM K S, HONG S M, HAN K, et al. Association of non-alcoholic fatty liver disease with cardiovascular disease and all cause death in patients with type 2 diabetes mellitus: nationwide population based study[J]. BMJ, 2024, 384: e076388. DOI:10.1136/bmj-2023-076388 . |
| 25 | SEBASTIAN S A, PADDA I, JOHAL G. Cardiovascular-kidney-metabolic (CKM) syndrome: a state-of-the-art review[J]. Curr Probl Cardiol, 2024, 49(2):102344. DOI:10.1016/j.cpcardiol. 2023.102344 . |
| 26 | WANG Y Q, ZHU J K, TANG Y, et al. Association between pulp and periapical disease with type 2 diabetes: a bidirectional Mendelian randomization[J]. Int Endod J, 2024, 57(5):566-575. DOI:10.1111/iej.14034 . |
| 27 | NADIA A, HASSAN A, MUSHRIQ A. Oral flora and functional dysbiosis of cleft lip and palate patients: a scoping review[J]. Spec Care Dent, 2023, 44(2):255-268. DOI:10.1111/scd.12872 . |
| 28 | XU J Y, YU L, YE S R, et al. Oral microbiota-host interaction: the chief culprit of alveolar bone resorption[J]. Front Immunol, 2024, 15:1254516. DOI:10.3389/fimmu.2024.1254516 . |
| 29 | 李宗泽, 赵泽, 张文菲, 等. 16S核糖体DNA Amplicon测序法分析2型糖尿病患者口腔微生物多样性[J]. 转化医学杂志, 2017, 6(3):151-156. DOI: 10.3969/j.issn.2095-3097.2017.03.006 . |
| LI Z Z, ZHAO Z, ZHANG W F, et al. Analysis of oral microbial diversity in patients with type 2 diabetes mellitus by 16S ribosome DNA Amplicon sequencing[J]. Transl Med J, 2017, 6(3):151-156. DOI: 10.3969/j.issn.2095-3097.2017.03.006 . | |
| 30 | 程梦蓉, 秦立果, 姜大昭, 等. β-羟基丁酸对糖尿病患者口腔菌群结构和丰度的调节作用[J]. 中国科技论文, 2020, 15(12):1377-1385. DOI: 10.3969/j.issn.2095-2783.2020.12.006 . |
| CHENG M R, QIN L G, JIANG D Z, et al. Modulatory effect of β-hydroxybutyrate on the structure and abundance of oral microbiota in diabetic populations[J]. China Sci, 2020, 15(12):1377-1385. DOI: 10.3969/j.issn.2095-2783.2020.12.006 . | |
| 31 | GAO C, GUO Y, CHEN F. Cross-cohort microbiome analysis of salivary biomarkers in patients with type 2 diabetes mellitus[J]. Front Cell Infect Microbiol, 2022, 12:816526. DOI: 10.3389/fcimb.2022.816526 . |
| 32 | 赵玉兰, 杨艺, 李进涛, 等. 2型糖尿病db/db小鼠骨强度和肠道内容物菌群的变化[J]. 中国比较医学杂志, 2023, 33(6):1-7. DOI: 10.3969/j.issn.1671-7856.2023.06.001 . |
| ZHAO Y L, YANG Y, LI J T, et al. Changes of bone strength and gut microbiota in type 2 diabetic db/db mice[J]. Chin J Comp Med, 2023, 33(6):1-7. DOI: 10.3969/j.issn.1671-7856.2023.06.001 . | |
| 33 | 朱洪杨, 刘烨, 李家荣, 等. 基于肠道菌群分析探讨葛根减轻T2DM db/db小鼠胰岛素抵抗的作用及机制[J]. 中国中药杂志, 2023, 48(17):4693-4701. DOI: 10.19540/j.cnki.cjcmm.20230418.402 . |
| ZHU H Y, LIU Y, LI J R, et al. Effect and mechanism of Puerariae Lobatae Radix in alleviating insulin resistance in T2DM db/db mice based on intestinal flora[J]. China J Chin Mater Med, 2023, 48(17):4693-4701. DOI: 10.19540/j.cnki.cjcmm.20230418.402 . | |
| 34 | 彭川, 胡学芳, 陈正涛, 等. 基于16S rRNA技术研究蒌连丸对2型糖尿病db/db小鼠肠道菌群的影响[J].中国实验方剂学杂志, 2023, 29(12): 63-70. DOI: 10.13422/j.cnki.syfjx.20222107 . |
| PENG C, HU X F, CHEN Z T, et al. Effect of Loulianwan on gut microbiota in db/db mice with type 2 diabetes mellitus based on 16S rRNA sequencing technology[J]. Chin J Exp Tradit Med Form, 2023, 29(12): 63-70. DOI: 10.13422/j.cnki.syfjx.20222107 . | |
| 35 | 刘军, 丁登峰, 高伟, 等. 应用16S rRNA基因测序比较分析不同品系1型糖尿病小鼠肠道菌群的异同[J]. 实验动物科学, 2023, 40(5):24-32. DOI: 10.3969/j.issn.1006-6179.2023.05.005 . |
| LIU J, DING D F, GAO W, et al. Use of 16S rRNA sequencing for comparative analysis of gut microbiome in different strains of type 1 diabetic mice[J]. Lab Anim Sci, 2023, 40(5):24-32. DOI: 10.3969/j.issn.1006-6179.2023.05.005 . | |
| 36 | AEMAIMANAN P, AMIMANAN P, TAWEECHAISUPAPONG S. Quantification of key periodontal pathogens in insulin-dependent type 2 diabetic and non-diabetic patients with generalized chronic periodontitis[J]. Anaerobe, 2013, 22:64-68. DOI:10.1016/j.anaerobe.2013.06.010 . |
| 37 | GU M, WANG P P, XIANG S K, et al. Effects of type 2 diabetes and metformin on salivary microbiota in patients with chronic periodontitis[J]. Microb Pathog, 2021, 161(Pt B):105277. DOI:10.1016/j.micpath.2021.105277 . |
| 38 | HE J, SHEN X, FU D, et al. Human periodontitis-associated salivary microbiome affects the immune response of diabetic mice[J]. J Oral Microbiol, 2022, 14(1):2107814. DOI:10.1080/20002297.2022.2107814 . |
| 39 | 李丽丽, 谢晓婷, 吴贇, 等. 牙周炎与糖尿病关联机制的研究进展[J]. 四川大学学报(医学版), 2023, 54(1):71-76.DOI: 10.12182/20230160203 . |
| LI L L, XIE X T, WU Y, et al. Advances in research on the mechanism of association between periodontitis and diabetes mellitus[J]. J Sichuan Univ (Med Sci), 2023, 54(1):71-76. DOI: 10.12182/20230160203 . |
| [1] | YANG Jin, YU Shiya, LIN Nan, FANG Yongchao, ZHAO Hu, QIU Jinwei, LIN Hongming, CHEN Huiyan, WANG Yu, WU Weihang. Effect of Modified Duodenal Exclusion Surgery on Glucose Metabolism in Rats with Type 2 Diabetes Mellitus [J]. Laboratory Animal and Comparative Medicine, 2024, 44(5): 523-530. |
| [2] | WANG Xiaoyu, JIANG Shengyao, LIN Zhibing, CUI Li. Effects of Diabetes on Colorectal Cancer/Breast Cancer Progression and Intestinal Flora Based on a Diabetes-tumor Mouse Model [J]. Laboratory Animal and Comparative Medicine, 2021, 41(6): 469-479. |
| [3] | JIANG Xia, QIAN Haojie, WEI Xun, ZHENG Yuxuan, ZHOU Zhengyu. Research Progress in Construction and Application of Diabetes Model in Zebrafish [J]. Laboratory Animal and Comparative Medicine, 2020, 40(6): 547-552. |
| [4] | HOU Wei, WAN Dun. Alleviation Effect of Formononetin on Osteoporosis in Type 2 Diabetes Mellitus Complicated with Diabetic Bone Disease#br# [J]. Laboratory Animal and Comparative Medicine, 2020, 40(3): 232-. |
| [5] | CHEN Zhi-ling, WU Hua, SONG Xin-lei. Investigation on Mouse Model of Type 2 Diabetes Mellitus Combined with Alzheimer's Disease [J]. Laboratory Animal and Comparative Medicine, 2018, 38(1): 29-35. |
| [6] | CHI Jun, GONG Hui, HE Yue-wei, WAN Ying-han, FEI Jian, KUANG Ying. Generation and Phenotyping Analysis of Lep Gene Knockout Mice [J]. Laboratory Animal and Comparative Medicine, 2017, 37(5): 344-351. |
| [7] | WANG Fei-fei, GONG Jing-jing, HUO Xue-yun, LU-jing, GUO-Meng, LIU-Xin, LI Chang-long, DU Xiao-yan, CHEN Zhen-wen, LV Jian-yi. Analysis on Cyclooxygenase 2 Expression in Different Tissues from Spontaneous Diabetic Mongolian Gerbils [J]. Laboratory Animal and Comparative Medicine, 2017, 37(2): 118-122. |
| [8] | LI Yin-yin, GONG Jing-jing, WU Shao-liang, LI Xiao-hong, WANG Cun-long, HUO Xue-yun, LU Jing, LV Jian-yi, LIU Xin, GUO Meng, LI Chang-long, CHEN Zhen-wen, DU Xiao-yan. Expression of ND3 in 5 Tissues of Hereditary Diabetic Mongolian Gerbils [J]. Laboratory Animal and Comparative Medicine, 2017, 37(1): 6-10. |
| [9] | LIANG Yong-jun, ZHANG Peng, WANG Yue-qian, GAO Li-li, CAO Ting, QIAO Zheng-dong. High-fat Diet Induced Type 2 Diabetes Mellitus and Obesity in C57BL/6 Mice [J]. Laboratory Animal and Comparative Medicine, 2016, 36(5): 361-364. |
| [10] | ZHENG Jin-hua, HE Min-zhu, XI Jun, ZHUANG Hua, WANG Zhu-gang, KUANG Ying. Generation of Lepr Gene Knockin Mouse Model and its Phenotypic Analysis [J]. Laboratory Animal and Comparative Medicine, 2014, 34(5): 345-352. |
| [11] | CHEN Li-Xin, ZHAO Xian-zhe, QIAO Wei-wei, GU Jian-Zhong, WANG Long-Xia. Study on Gut Hormones in GK Rats after Different Length Duodenum-Preserving Duodenal-Jejunal Bypass Surgery [J]. Laboratory Animal and Comparative Medicine, 2014, 34(3): 223-227. |
| [12] | WANG Ling-yun, WANG Ping, SUN Yu-xiu, LI Dan-dan, LIU Song, ZHANG Qiu, LU Yun-xia. Development and Evaluation of a Modeling Method For Experimental Rat with Type 2 Diabetes Mellitus [J]. Laboratory Animal and Comparative Medicine, 2011, 31(3): 160-165. |
| [13] | ZHAO Xian-zhe, QIAO Wei-wei. The Research Advancement on Etiopathology of Goto-Kakizaki Diabetic Rat [J]. Laboratory Animal and Comparative Medicine, 2011, 31(2): 140-146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||