
Laboratory Animal and Comparative Medicine ›› 2023, Vol. 43 ›› Issue (1): 11-20.DOI: 10.12300/j.issn.1674-5817.2022.100
• Animal Model of Human Disease: Pharmacology • Previous Articles Next Articles
Ling YANG1( ), Di ZHUANG2, Lilun JIN1(
), Di ZHUANG2, Lilun JIN1( )(
)( )
)
Received:2022-07-04
Revised:2022-10-02
Online:2023-02-25
Published:2023-03-09
Contact:
Lilun JIN
CLC Number:
Ling YANG, Di ZHUANG, Lilun JIN. Screening of Differentially Expressed Genes in Rat Synovitis by Transcriptome Sequencing and in Vitro Verification of Therapeutic Target of Fraxetin[J]. Laboratory Animal and Comparative Medicine, 2023, 43(1): 11-20.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2022.100
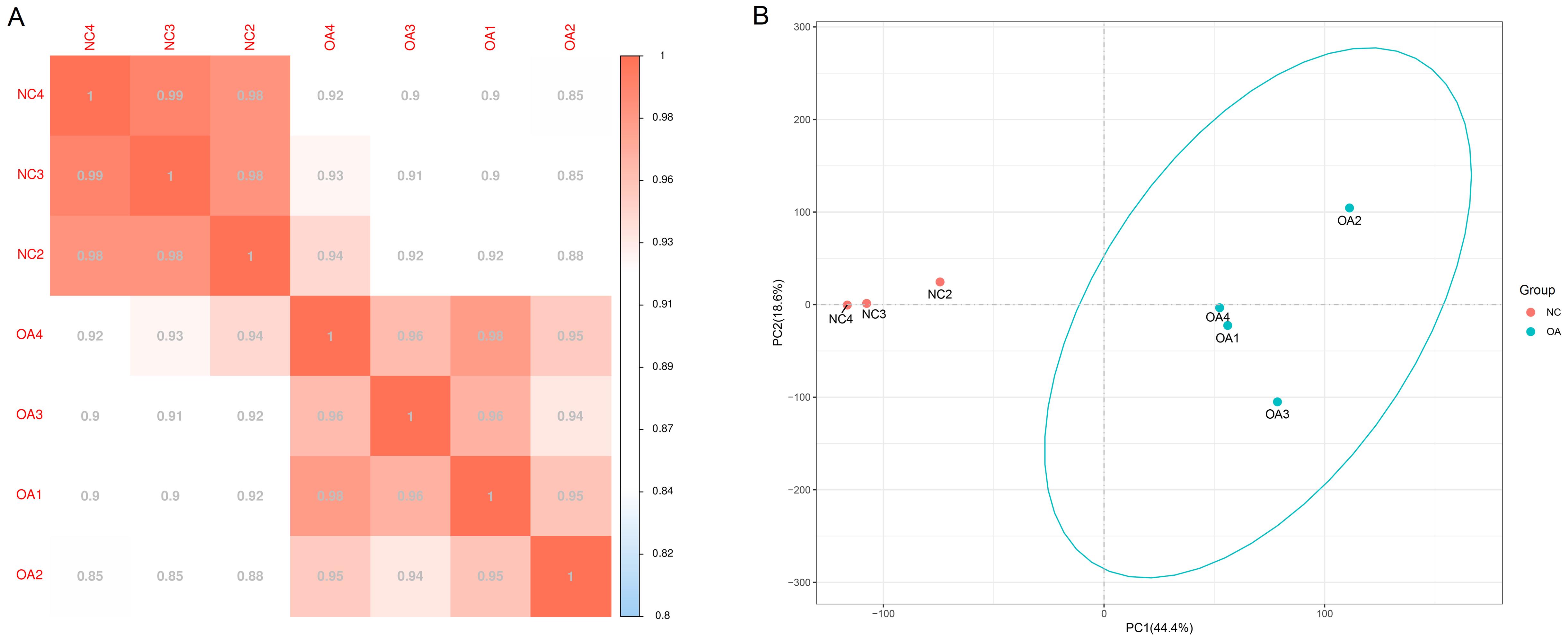
Figure 1 Gene expression correlation analysis (A) and principal components analysis (B) between the samplesNote: A, showed the correlation test, the closer the correlation coefficient is to 1, the higher the sample similarity is. In the negative control group (NC) and knee osteoarthritis model group (OA), the intra-group samples are more similar than inter-group samples. B, performed a principal component (PC) analysis, samples from the same group packed together in the figure, which indicates that the sequencing data are of reliable quality.
| 差异Difference | 基因Gene | log2FC | P值P value | 校正P值Padj value |
|---|---|---|---|---|
| 上调Up-regulated | Mmp12 | 9.933 625 864 | 9.132 91×10-11 | 3.537 83×10-9 |
| Acod1 | 9.272 106 280 | 0.000 127 507 | 0.000 802 842 | |
| Col2a1 | 9.153 847 197 | 7.690 69×10-9 | 1.807 62×10-7 | |
| Atp6v0d2 | 8.282 894 807 | 5.603 80×10-6 | 5.607 99×10-5 | |
| Fbn2 | 7.973 227 345 | 2.921 38×10-37 | 3.387 43×10-34 | |
| Acan | 7.718 643 189 | 4.203 11×10-20 | 1.015 34×10-17 | |
| Ocstamp | 7.591 893 578 | 0.001 876 497 | 0.007 726 778 | |
| LOC103690326 | 7.546 217 455 | 7.903 40×10-5 | 0.000 529 726 | |
| Cxcl2 | 7.422 908 965 | 1.713 86×10-5 | 0.000 145 198 | |
| Mmp9 | 7.188 248 072 | 2.374 04×10-11 | 1.027 150×10-9 | |
| 下调Down-regulated | Plekha2 | -1.000 555 825 | 1.696 39×10-5 | 0.000 143 999 |
| Nudt7 | -1.001 123 965 | 5.602 09×10-5 | 0.000 395 268 | |
| Cdc42bpg | -1.002 206 901 | 0.000 106 010 | 0.000 684 033 | |
| Mtus1 | -1.002 219 197 | 0.000 139 053 | 0.000 863 768 | |
| Ppp1r37 | -1.002 289 468 | 4.948 870×10-20 | 1.179 120×10-17 | |
| Oard1 | -1.003 517 453 | 0.000 375 535 | 0.002 019 066 |
Table 1 Genes up-regulated and down-regulated in synovial tissues of knee osteoarthritis model rats iduced by monosodium iodoacetate
| 差异Difference | 基因Gene | log2FC | P值P value | 校正P值Padj value |
|---|---|---|---|---|
| 上调Up-regulated | Mmp12 | 9.933 625 864 | 9.132 91×10-11 | 3.537 83×10-9 |
| Acod1 | 9.272 106 280 | 0.000 127 507 | 0.000 802 842 | |
| Col2a1 | 9.153 847 197 | 7.690 69×10-9 | 1.807 62×10-7 | |
| Atp6v0d2 | 8.282 894 807 | 5.603 80×10-6 | 5.607 99×10-5 | |
| Fbn2 | 7.973 227 345 | 2.921 38×10-37 | 3.387 43×10-34 | |
| Acan | 7.718 643 189 | 4.203 11×10-20 | 1.015 34×10-17 | |
| Ocstamp | 7.591 893 578 | 0.001 876 497 | 0.007 726 778 | |
| LOC103690326 | 7.546 217 455 | 7.903 40×10-5 | 0.000 529 726 | |
| Cxcl2 | 7.422 908 965 | 1.713 86×10-5 | 0.000 145 198 | |
| Mmp9 | 7.188 248 072 | 2.374 04×10-11 | 1.027 150×10-9 | |
| 下调Down-regulated | Plekha2 | -1.000 555 825 | 1.696 39×10-5 | 0.000 143 999 |
| Nudt7 | -1.001 123 965 | 5.602 09×10-5 | 0.000 395 268 | |
| Cdc42bpg | -1.002 206 901 | 0.000 106 010 | 0.000 684 033 | |
| Mtus1 | -1.002 219 197 | 0.000 139 053 | 0.000 863 768 | |
| Ppp1r37 | -1.002 289 468 | 4.948 870×10-20 | 1.179 120×10-17 | |
| Oard1 | -1.003 517 453 | 0.000 375 535 | 0.002 019 066 |
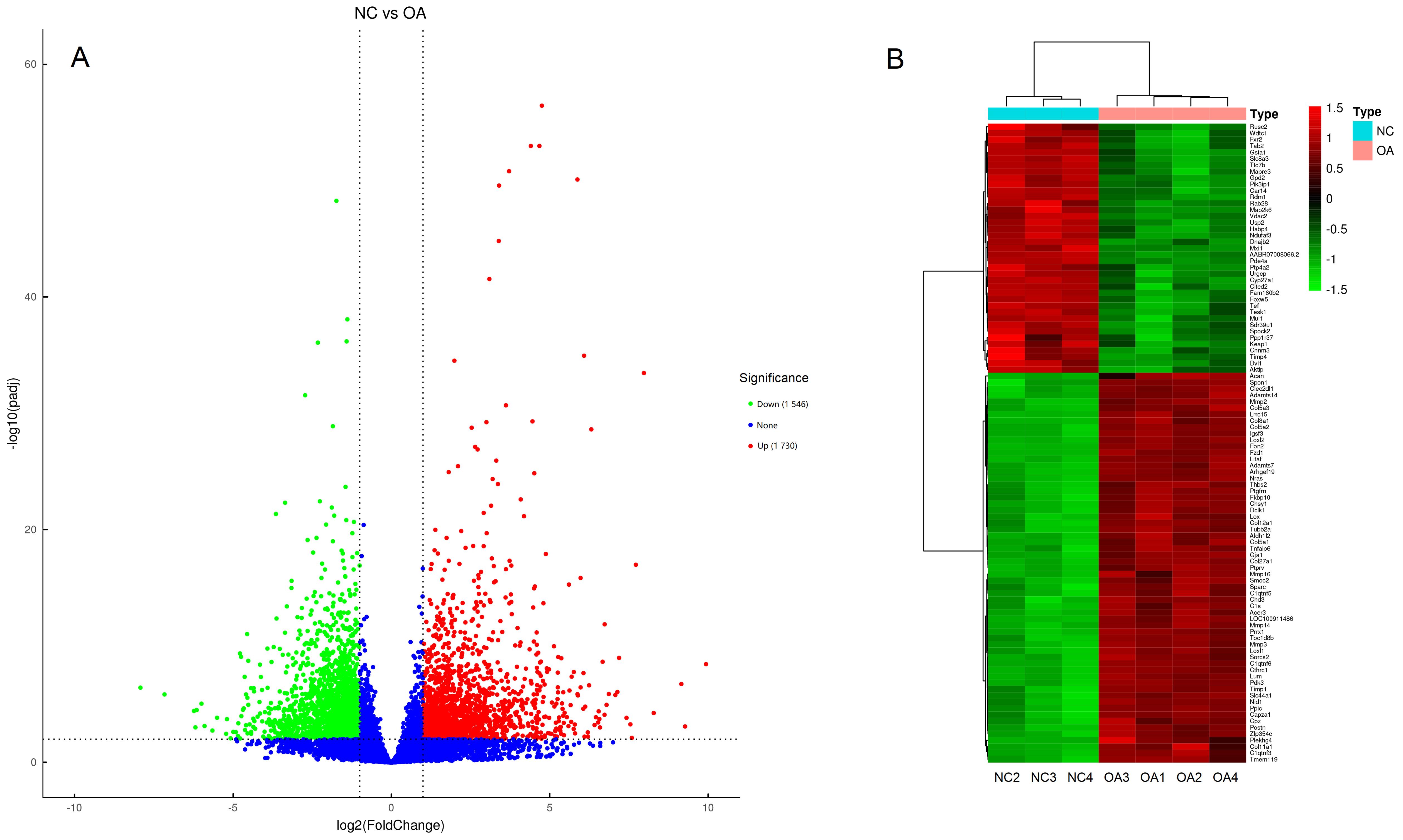
Figure 2 Volcano plots (A) and heat map (B) of differentially expressed genesNote:A, the abscissa is log2FC, the ordinate is -log10(adjusted P). Red or green dots indicate significantly up-regulated and down-regulated genes, respectively. While, genes with no significantly differentially expressed are indicated with blue dots. B, Gene heat map, the abscissa is sample names, the ordinate is gene names, red indicates high gene expression, while green indicates low expression. NC, negative control group; OA, knee osteoarthritis model group.
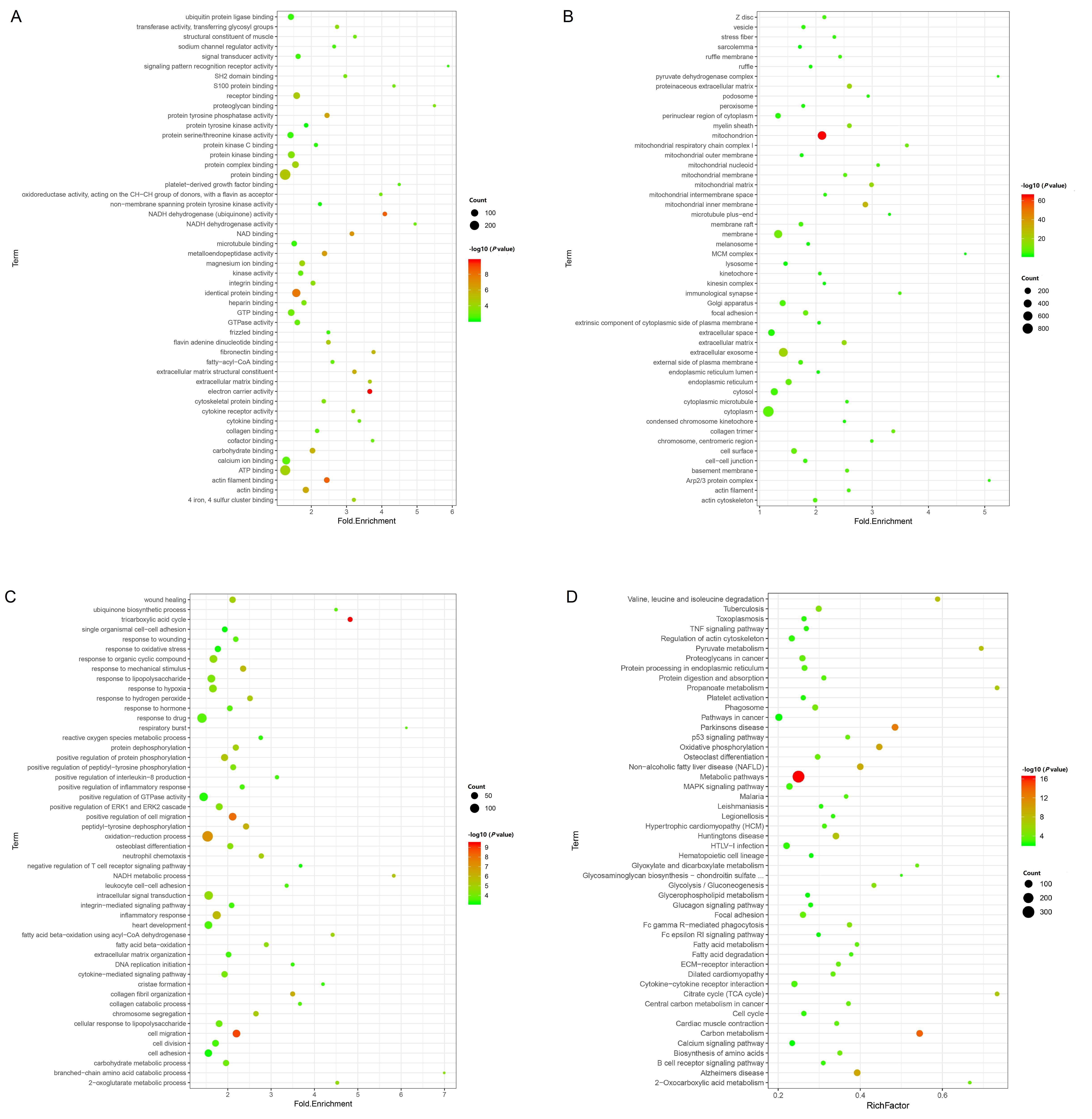
Figure 3 Gene ontology (GO) enrichment analysis (A-C) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (D) of differentially expressed genesNote:A-C,GO enrichment analysis showed three ontologies “molecular function”(A), “cellular component”(B) and “biological process”(C). The abscissa is Fold Enrichment, the ordinate is term, the size of the bubbles indicates the number of term-enriched genes. The bigger the bubble is, the more genes enriched under the term. The colors of different bubbles correspond to P-value, the increasingly reddish colors means the more significant of P-value.
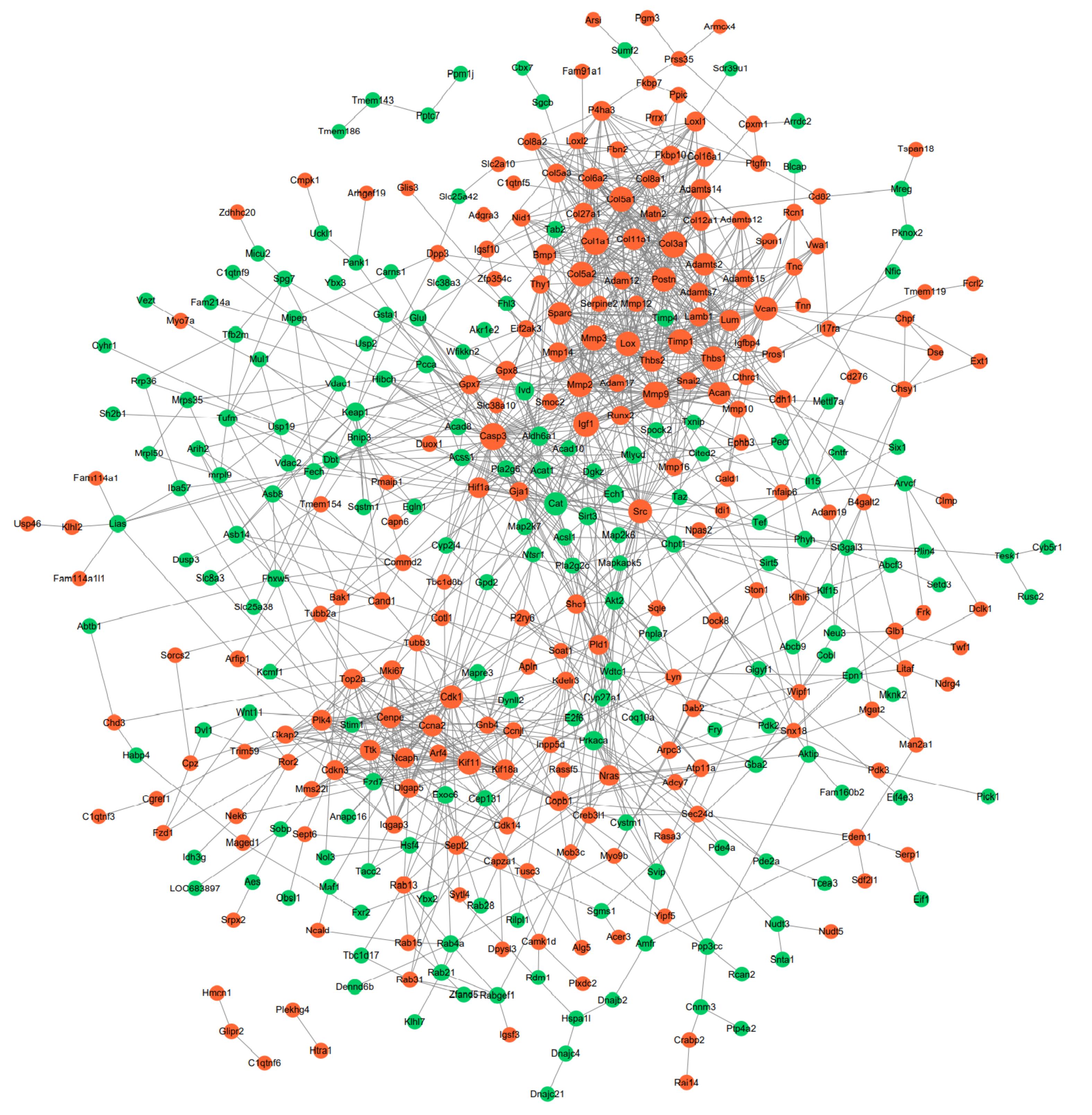
Figure 4 The protein-protein interaction (PPI) networks of differentially expressed genesNote:The size of the points means the degree of the node. The more protein connects with the node, the bigger the node will become. The red notes mean the up-regulated genes, the green ones mean down-regulated genes.
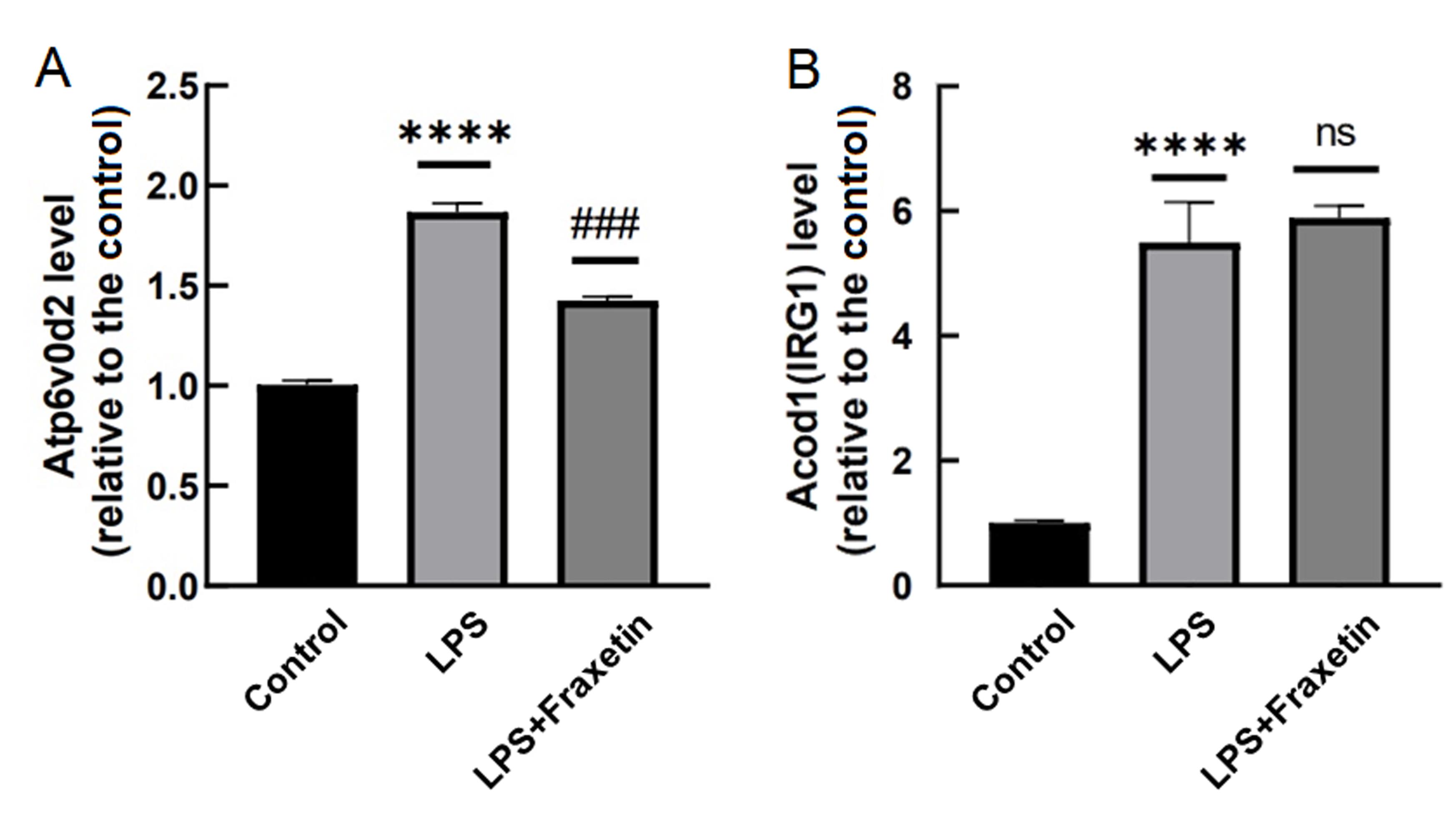
Figure 5 Atp6v0d2 and Acod1 gene expression in synovial macrophage RAW264.7 cells treated with lipopolysaccharide (LPS) and Fraxetin detected by real-time fluorescence quantitative PCRNote:Compared with the control group,****P<0.000 1; Compared with the LPS group, ###P<0.001. ns indicated none statistically significant differences. n=3。
| 1 | VINA E R, KWOH C K. Epidemiology of osteoarthritis: literature update[J]. Curr Opin Rheumatol, 2018, 30(2):160-167. DOI: 10.1097/BOR.0000000000000479 . |
| 2 | ABRAMOFF B, CALDERA F E. Osteoarthritis: pathology, diagnosis, and treatment options[J]. Med Clin North Am, 2020, 104(2):293-311. DOI: 10.1016/j.mcna.2019.10.007 . |
| 3 | EYMARD F, PIGENET A, CITADELLE D, et al. Knee and hip intra-articular adipose tissues (IAATs) compared with autologous subcutaneous adipose tissue: a specific phenotype for a central player in osteoarthritis[J]. Ann Rheum Dis, 2017, 76(6):1142-1148. DOI: 10.1136/annrheumdis-2016-210478 . |
| 4 | NEOGI T, GUERMAZI A, ROEMER F, et al. Association of joint inflammation with pain sensitization in knee osteoarthritis: the multicenter osteoarthritis study[J]. Arthritis Rheumatol, 2016, 68(3):654-661. DOI: 10.1002/art.39488 . |
| 5 | KEMBLE S, CROFT A P. Critical role of synovial tissue-resident macrophage and fibroblast subsets in the persistence of joint inflammation[J]. Front Immunol, 2021, 12:715894. DOI: 10.3389/fimmu.2021.715894 . |
| 6 | LOPES E B P, FILIBERTI A, HUSAIN S ALI, et al. Immune contributions to osteoarthritis[J]. Curr Osteoporos Rep, 2017, 15(6):593-600. DOI: 10.1007/s11914-017-0411-y . |
| 7 | 马振源, 莫蕙. 中医贴敷治疗膝骨关节炎的研究进展[J]. 南京中医药大学学报, 2020, 36(6):926-930. DOI: 10.14148/j.issn.1672-0482.2020.0926 . |
| MA Z Y, MO H. Research progress of traditional Chinese medicine sticking therapy in the treatment of knee osteoarthritis[J]. J Nanjing Univ Tradit Chin Med, 2020, 36(6):926-930. DOI: 10.14148/j.issn.1672-0482.2020.0926 . | |
| 8 | 侯成志, 李秋月, 魏戌, 等. 独活寄生汤治疗膝骨关节炎的研究[J]. 中国中医基础医学杂志, 2021, 27(11):1843-1846. DOI: 10.19945/j.cnki.issn.1006-3250.2021.11.036 . |
| HOU C Z, LI Q Y, WEI X, et al. Study on Duhuo Jisheng Decoction in the treatment of knee osteoarthritis[J]. J Basic Chin Med, 2021, 27(11):1843-1846. DOI: 10.19945/j.cnki.issn.1006-3250.2021.11.036 . | |
| 9 | 王倩, 杨凯, 马强, 等. 中医药治疗膝骨关节炎研究进展[J]. 陕西中医药大学学报, 2022, 45(3):130-134. DOI: 10.13424/j.cnki.jsctcm.2022.03.029 . |
| WANG Q, YANG K, MA Q, et al. Research progress on treatment of knee osteoarthritis with traditional Chinese medicine[J]. J Shaanxi Univ Chin Med, 2022, 45(3):130-134. DOI: 10.13424/j.cnki.jsctcm.2022.03.029 . | |
| 10 | HUSSAIN S M, NEILLY D W, BALIGA S, et al. Knee osteoarthritis: a review of management options[J]. Scott Med J, 2016, 61(1):7-16. DOI: 10.1177/0036933015619588 . |
| 11 | WANG Q, ZHUANG D, FENG W, et al. Fraxetin inhibits interleukin-1β-induced apoptosis, inflammation, and matrix degradation in chondrocytes and protects rat cartilage in vivo [J]. Saudi Pharm J, 2020, 28(12):1499-1506. DOI: 10.1016/j.jsps.2020.09.016 . |
| 12 | 金灵璐, 朱尉东, 仲卫红, 等. 升麻素苷治疗膝骨关节炎的作用机制研究[J]. 风湿病与关节炎, 2021, 10(12):45-48. |
| JIN L L, ZHU W D, ZHONG W H, et al. On the mechanism of cimicifugin in the treatment of knee osteoarthritis[J]. Rheum Arthritis, 2021, 10(12):45-48. | |
| 13 | 高嘉美, 潘赐明, 郗域江, 等. 当归治疗骨关节炎作用机制的研究进展[J]. 风湿病与关节炎, 2021, 10(11):77-80. |
| GAO J M, PAN C M, XI Y J, et al. Research progress on the mechanism of Angelica sinensis in the treatment of osteoarthritis[J]. Rheum Arthritis, 2021, 10(11):77-80. | |
| 14 | DESHMUKH V, O'GREEN A L, BOSSARD C, et al. Modulation of the Wnt pathway through inhibition of CLK2 and DYRK1A by lorecivivint as a novel, potentially disease-modifying approach for knee osteoarthritis treatment[J]. Osteoarthritis Cartilage, 2019, 27(9): 1347-1360. DOI: 10.1016/j.joca.2019. 05. 006 . |
| 15 | LI Z H, HUANG Z Y, ZHANG H, et al. P2X7 receptor induces pyroptotic inflammation and cartilage degradation in osteoarthritis via NF-κB/NLRP3 crosstalk[J]. Oxidative Med Cell Longev, 2021, 2021:1-16. DOI: 10.1155/2021/8868361 . |
| 16 | ZHENG L L, ZHANG Z J, SHENG P Y, et al. The role of metabolism in chondrocyte dysfunction and the progression of osteoarthritis[J]. Ageing Res Rev, 2021, 66: 101249. DOI: 10.1016/j.arr.2020.101249 . |
| 17 | MOBASHERI A, RAYMAN M P, GUALILLO O, et al. The role of metabolism in the pathogenesis of osteoarthritis[J]. Nat Rev Rheumatol, 2017, 13(5):302-311. DOI: 10.1038/nrrheum.2017.50 . |
| 18 | 朱璐丹. 秦皮素对嗜水气单胞菌的抗感染机制研究[D].上海:上海海洋大学, 2020. DOI: 10.27314 /d.cnki.gsscu. 2020.000028 . |
| ZHU L D. Anti-infective mechanism of fraxetin against aeromonas hydrophila[D]. Shanghai: Shanghai Ocean University, 2020. DOI: 10.27314 /d.cnki.gsscu. 2020.000028 . | |
| 19 | 曹桂云, 宁波, 秦金淼, 等. 基于标准汤剂的秦皮(白蜡树)配方颗粒质量标准分析[J/OL]. 中国实验方剂学杂志:1-11(2022-11-22)[2023-02-22]. . |
| CAO G Y, NING B, QIN J M, et al. Quality standard analysis of fraxinus fraxinus dispensing granules based on standard decoction[J/OL]. Chin J Exp Formul: 1-11(2022-11-22)[2023-02-22]. . | |
| 21 | WU R, CHEN F, WANG N, et al. ACOD1 in immunometabolism and disease[J]. Cell Mol Immunol, 2020, 17(8):822-833. DOI: 10.1038/s41423-020-0489-5 . |
| 22 | GRIFFIN T M, SCANZELLO C R. Innate inflammation and synovial macrophages in osteoarthritis pathophysiology[J]. Clin Exp Rheumatol, 2019, 37 Suppl 120(5):57-63. |
| 23 | RU L B. Synovium and the innate inflammatory network in osteoarthritis progression[J]. Curr Rheumatol Rep, 2013, 15(5):323. DOI: 10.1007/s11926-013-0323-5 . |
| 24 | XIA Y, LIU N, XIE X X, et al. The macrophage-specific V-ATPase subunit ATP6V0D2 restricts inflammasome activation and bacterial infection by facilitating autophagosome-lysosome fusion[J]. Autophagy, 2019, 15(6):960-975. DOI: 10.1080/15548627.2019.1569916 . |
| [1] | Ruolin CUI, Qing WANG, Ling YANG, Wenchang FENG, Zhiwei LIU, Weiran TENG, Bitao MA, Leyang WANG, Liping QIN, Lilun JIN. Repair Effects of Xiaoyusan New Formula on Cartilage Injury and MMP-13 Expression in Knee Osteoarthritis Model Rabbits [J]. Laboratory Animal and Comparative Medicine, 2023, 43(1): 30-38. |
| [2] | XIA Mengxiong, HAN Haihui, LIANG Qianqian, ZHAI Weitao, XU Hao. Research Progress on Animal Models of Osteoarthritis [J]. Laboratory Animal and Comparative Medicine, 2020, 40(2): 159-165. |
| [3] | XIAO Kunlin, ZHANG Rui, Sun Hong, XIAO Kuntai, MA Jianbing. Evaluation of Monoiodoacetic Acid-induced Knee Osteoarthritis SD Rats with Diseasse Progression at Different Time Points [J]. Laboratory Animal and Comparative Medicine, 2020, 40(1): 47-52. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||