
Laboratory Animal and Comparative Medicine ›› 2023, Vol. 43 ›› Issue (5): 548-558.DOI: 10.12300/j.issn.1674-5817.2023.073
• Research Reports • Previous Articles Next Articles
Liya ZHAO1,2( ), Liju NI1(
), Liju NI1( ), Caiqin ZHANG3, Jianping TANG1,2, Yangzheng YAO4, Yanyan NIE1, Xiaoxue GU2, Ying ZHAO1(
), Caiqin ZHANG3, Jianping TANG1,2, Yangzheng YAO4, Yanyan NIE1, Xiaoxue GU2, Ying ZHAO1( )(
)( )
)
Received:2023-06-09
Revised:2023-08-08
Online:2023-10-25
Published:2023-10-25
Contact:
Ying ZHAO
CLC Number:
Liya ZHAO,Liju NI,Caiqin ZHANG,et al. Establishing a Genetic Detection Protocol of Single Nucleotide Polymorphisms Panels in Inbred Rats Based on Multiplex PCR-LDR[J]. Laboratory Animal and Comparative Medicine, 2023, 43(5): 548-558. DOI: 10.12300/j.issn.1674-5817.2023.073.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2023.073
序号 No. | SNP位点 Reference SNP ID | 染色体 Chromosome | 引物名称 Primer name | 引物序列 Primer sequences |
|---|---|---|---|---|
| a1 | rs65970293 | chr1 | rs65970293-up | TGCACATTGGGACTGCTTTA |
| rs65970293-low | ATCTGGCATGGACAGAGCTT | |||
| a2 | rs106996961 | chr2 | rs106996961-up | ATAGGGCCAAGGTCCCATTC |
| rs106996961-low | GAAAAGAACGCTAATCACAGGACA | |||
| a3 | rs63860341 | chr3 | rs63860341-up | CAATGAATCAAAGGCAACCA |
| rs63860341-low | CCACGGTCCCTATGTGAACT | |||
| a4 | rs64750237 | chr4 | rs64750237-up | CAGGTATGGCTTTGGAGGAA |
| rs64750237-low | GTTTTCAGCTGGAGCTGTCC | |||
| a5 | rs107261116 | chr5 | rs107261116-up | CAGCATTGGCAAGTCTTCCT |
| rs107261116-low | TGATAAAAGGAGCAGCAGCA | |||
| a6 | rs64102787 | chr6 | rs64102787-up | CGGGGATCAAAGAGACAAAA |
| rs64102787-low | ATAGTGCCAAAGCCCAACAC | |||
| a7 | rs105187832 | chr7 | rs105187832-up | AGACTACTGGGCTGCTCACC |
| rs105187832-low | CATGGCACACCTTTCTTTCA | |||
| a8 | rs64682441 | chr9 | rs64682441-up | GCTGGAAGTCTCTGGAGTGG |
| rs64682441-low | ACAAGTGGACTGGCTCAGGT | |||
| a9 | rs65067166 | chr10 | rs65067166-up | ATCCCTCTCTTTGCCAGGAT |
| rs65067166-low | GTAGGGTGAGGCTGCTTCTG | |||
| a10 | rs8167985 | chr11 | rs8167985-up | ACCCCCTCTTGCTTTCTGTT |
| rs8167985-low | CTGCTCTTGGATGAAAACCA | |||
| b1 | rs106672258 | chr12 | rs106672258-up | GCCCTTCCCAAGGTCATACT |
| rs106672258-low | AAAGCACAAAAATGCCATCC | |||
| b2 | rs65921305 | chr13 | rs65921305-up | AACCTGGTGATTTGTCAGAGC |
| rs65921305-low | GGCAGACTCCAGGAAGATTG | |||
| b3 | rs106458396 | chr15 | rs106458396-up | AGAGCAGCTGCCAAAATAGC |
| rs106458396-low | AAACCTGGCTTTAGCAAGCA | |||
| b4 | rs106888236 | chr16 | rs106888236-up | TGTGAAGAACACAGGCTCTCC |
| rs106888236-low | TGCCAAACACCATGTAATGAA | |||
| b5 | rs63814610 | chr17 | rs63814610-up | TAGGACAGGGGGAACAACTG |
| rs63814610-low | CCCAAGCATCCTTGACTGTT | |||
| b6 | rs8152541 | chr18 | rs8152541-up | CGTGTCATTCCTGACTGTGG |
| rs8152541-low | ACAGGAGCTGGCACTGAATC | |||
| b7 | rs105824121 | chr19 | rs105824121-up | TTCTGCAATCAAGGGTGGAT |
| rs105824121-low | GGCCACCAAAGATACACAGG | |||
| b8 | rs13453252 | chr20 | rs13453252-up | TACGCCATTTTGCGACATTA |
| rs13453252-low | GGTTCCCTACGAGGAACGAC | |||
| b9 | rs65108991 | chrX | rs65108991-up | CATCCCAGAAGCCCTGTTTA |
| rs65108991-low | GAAGTCAGTGCTGTGCCTCA | |||
| c1 | rs106891788 | chr1 | rs106891788-up | TCCATCTGCTTGGCCCTAAT |
| rs106891788-low | GTTGCAACCCACAGGTTGAG | |||
| c2 | rs107493604 | chr2 | rs107493604-up | TCCTCCTGGAAGACAGCAAC |
| rs107493604-low | GTTCCTGAGAAGGGGCTCTC | |||
| c3 | rs65632519 | chr3 | rs65632519-up | CCCTGTTTCAGCCTTTTGTG |
| rs65632519-low | GAGGAAGAGGGAAGGGAGAA | |||
| c4 | rs107163178 | chr4 | rs107163178-up | TTCTCCCACCGGAGACTTAG |
| rs107163178-low | GAGAGGGCATATGTGGGATG | |||
| c5 | rs105170847 | chr5 | rs105170847-up | TGGTTGGATGAATGGAGACA |
| rs105170847-low | CCTCAGGAGCACCAACATTT | |||
| c6 | rs106547562 | chr6 | rs106547562-up | GGTTTTGTCCTCAGCAGGTT |
| rs106547562-low | CTCTTAGGCATCTGGGGAAA | |||
| c7 | rs65502317 | chr7 | rs65502317-up | TGTGCATGCTTACAGTGGCTA |
| rs65502317-low | AGCCAACTGGAGCCACATAG | |||
| c8 | rs105694545 | chr8 | rs105694545-up | TCAGCAGTGCTCAGCAGTTT |
| rs105694545-low | CGTAACATCATTTGGCATCG | |||
| c9 | rs65801920 | chr9 | rs65801920-up | TGGCTCTTGGTCATCATCTG |
| rs65801920-low | TTTTGCAGCCTGTGTGTAGC | |||
| c10 | rs105168947 | chr10 | rs105168947-up | GGCGGACAGAACATAGGTGT |
| rs105168947-low | CAGCCAAACTCCCAGATCAT | |||
| c11 | rs106880606 | chr11 | rs106880606-up | AGTGGGTTTAGCAAGCAGGA |
| rs106880606-low | GTCGTCGAAATGCAGGAGAT | |||
| d1 | rs8158863 | chr12 | rs8158863-up | GGTCACAGCCATTCTGGATT |
| rs8158863-low | GCCCTGAATGCAGGTGTAGT | |||
| d2 | rs105965886 | chr13 | rs105965886-up | CACCAGCAAAAGGGAAAAAG |
| rs105965886-low | CAGAACACAGCCTCAGTTGG | |||
| d3 | rs63919332 | chr14 | rs63919332-up | GTCCTGTCGCCCAATATCAC |
| rs63919332-low | ACAACAATGGCAGTGGTCAA | |||
| d4 | rs65809306 | chr15 | rs65809306-up | AGCACTTGTGTGGGAAGCTC |
| rs65809306-low | CCTGACAAAACCACATGCAG | |||
| d5 | rs65087257 | chr16 | rs65087257-up | CCTTCCTTTTCCACTGTCCA |
| rs65087257-low | CTTAGGGCCTAGGGGCTTTA | |||
| d6 | rs106021355 | chr17 | rs106021355-up | GGTGGTTTATGGCCAGGATA |
| rs106021355-low | AACTGGAGAGGAAAAGGGACA | |||
| d7 | rs106171441 | chr18 | rs106171441-up | TCAATTCTTGGCAGCATCAG |
| rs106171441-low | CTGGCCACTTCTCTGAGTCC | |||
| d8 | rs106777713 | chr19 | rs106777713-up | GTGAAGCGACATCGTCAAGA |
| rs106777713-low | CTTGGCTGTGAGATCGAGGT | |||
| d9 | rs104937988 | chr20 | rs104937988-up | GCTGCACAGAACTGATTCCA |
| rs104937988-low | TGCAAGAATTAAATAAAATCCAAA | |||
| d10 | rs105294203 | chrX | rs105294203-up | ACTCCCCAGAGGATTGTCG |
| rs105294203-low | ATGTTCAAGTGCAACGATGG |
Table 1 40 SNP loci and primer sequences
序号 No. | SNP位点 Reference SNP ID | 染色体 Chromosome | 引物名称 Primer name | 引物序列 Primer sequences |
|---|---|---|---|---|
| a1 | rs65970293 | chr1 | rs65970293-up | TGCACATTGGGACTGCTTTA |
| rs65970293-low | ATCTGGCATGGACAGAGCTT | |||
| a2 | rs106996961 | chr2 | rs106996961-up | ATAGGGCCAAGGTCCCATTC |
| rs106996961-low | GAAAAGAACGCTAATCACAGGACA | |||
| a3 | rs63860341 | chr3 | rs63860341-up | CAATGAATCAAAGGCAACCA |
| rs63860341-low | CCACGGTCCCTATGTGAACT | |||
| a4 | rs64750237 | chr4 | rs64750237-up | CAGGTATGGCTTTGGAGGAA |
| rs64750237-low | GTTTTCAGCTGGAGCTGTCC | |||
| a5 | rs107261116 | chr5 | rs107261116-up | CAGCATTGGCAAGTCTTCCT |
| rs107261116-low | TGATAAAAGGAGCAGCAGCA | |||
| a6 | rs64102787 | chr6 | rs64102787-up | CGGGGATCAAAGAGACAAAA |
| rs64102787-low | ATAGTGCCAAAGCCCAACAC | |||
| a7 | rs105187832 | chr7 | rs105187832-up | AGACTACTGGGCTGCTCACC |
| rs105187832-low | CATGGCACACCTTTCTTTCA | |||
| a8 | rs64682441 | chr9 | rs64682441-up | GCTGGAAGTCTCTGGAGTGG |
| rs64682441-low | ACAAGTGGACTGGCTCAGGT | |||
| a9 | rs65067166 | chr10 | rs65067166-up | ATCCCTCTCTTTGCCAGGAT |
| rs65067166-low | GTAGGGTGAGGCTGCTTCTG | |||
| a10 | rs8167985 | chr11 | rs8167985-up | ACCCCCTCTTGCTTTCTGTT |
| rs8167985-low | CTGCTCTTGGATGAAAACCA | |||
| b1 | rs106672258 | chr12 | rs106672258-up | GCCCTTCCCAAGGTCATACT |
| rs106672258-low | AAAGCACAAAAATGCCATCC | |||
| b2 | rs65921305 | chr13 | rs65921305-up | AACCTGGTGATTTGTCAGAGC |
| rs65921305-low | GGCAGACTCCAGGAAGATTG | |||
| b3 | rs106458396 | chr15 | rs106458396-up | AGAGCAGCTGCCAAAATAGC |
| rs106458396-low | AAACCTGGCTTTAGCAAGCA | |||
| b4 | rs106888236 | chr16 | rs106888236-up | TGTGAAGAACACAGGCTCTCC |
| rs106888236-low | TGCCAAACACCATGTAATGAA | |||
| b5 | rs63814610 | chr17 | rs63814610-up | TAGGACAGGGGGAACAACTG |
| rs63814610-low | CCCAAGCATCCTTGACTGTT | |||
| b6 | rs8152541 | chr18 | rs8152541-up | CGTGTCATTCCTGACTGTGG |
| rs8152541-low | ACAGGAGCTGGCACTGAATC | |||
| b7 | rs105824121 | chr19 | rs105824121-up | TTCTGCAATCAAGGGTGGAT |
| rs105824121-low | GGCCACCAAAGATACACAGG | |||
| b8 | rs13453252 | chr20 | rs13453252-up | TACGCCATTTTGCGACATTA |
| rs13453252-low | GGTTCCCTACGAGGAACGAC | |||
| b9 | rs65108991 | chrX | rs65108991-up | CATCCCAGAAGCCCTGTTTA |
| rs65108991-low | GAAGTCAGTGCTGTGCCTCA | |||
| c1 | rs106891788 | chr1 | rs106891788-up | TCCATCTGCTTGGCCCTAAT |
| rs106891788-low | GTTGCAACCCACAGGTTGAG | |||
| c2 | rs107493604 | chr2 | rs107493604-up | TCCTCCTGGAAGACAGCAAC |
| rs107493604-low | GTTCCTGAGAAGGGGCTCTC | |||
| c3 | rs65632519 | chr3 | rs65632519-up | CCCTGTTTCAGCCTTTTGTG |
| rs65632519-low | GAGGAAGAGGGAAGGGAGAA | |||
| c4 | rs107163178 | chr4 | rs107163178-up | TTCTCCCACCGGAGACTTAG |
| rs107163178-low | GAGAGGGCATATGTGGGATG | |||
| c5 | rs105170847 | chr5 | rs105170847-up | TGGTTGGATGAATGGAGACA |
| rs105170847-low | CCTCAGGAGCACCAACATTT | |||
| c6 | rs106547562 | chr6 | rs106547562-up | GGTTTTGTCCTCAGCAGGTT |
| rs106547562-low | CTCTTAGGCATCTGGGGAAA | |||
| c7 | rs65502317 | chr7 | rs65502317-up | TGTGCATGCTTACAGTGGCTA |
| rs65502317-low | AGCCAACTGGAGCCACATAG | |||
| c8 | rs105694545 | chr8 | rs105694545-up | TCAGCAGTGCTCAGCAGTTT |
| rs105694545-low | CGTAACATCATTTGGCATCG | |||
| c9 | rs65801920 | chr9 | rs65801920-up | TGGCTCTTGGTCATCATCTG |
| rs65801920-low | TTTTGCAGCCTGTGTGTAGC | |||
| c10 | rs105168947 | chr10 | rs105168947-up | GGCGGACAGAACATAGGTGT |
| rs105168947-low | CAGCCAAACTCCCAGATCAT | |||
| c11 | rs106880606 | chr11 | rs106880606-up | AGTGGGTTTAGCAAGCAGGA |
| rs106880606-low | GTCGTCGAAATGCAGGAGAT | |||
| d1 | rs8158863 | chr12 | rs8158863-up | GGTCACAGCCATTCTGGATT |
| rs8158863-low | GCCCTGAATGCAGGTGTAGT | |||
| d2 | rs105965886 | chr13 | rs105965886-up | CACCAGCAAAAGGGAAAAAG |
| rs105965886-low | CAGAACACAGCCTCAGTTGG | |||
| d3 | rs63919332 | chr14 | rs63919332-up | GTCCTGTCGCCCAATATCAC |
| rs63919332-low | ACAACAATGGCAGTGGTCAA | |||
| d4 | rs65809306 | chr15 | rs65809306-up | AGCACTTGTGTGGGAAGCTC |
| rs65809306-low | CCTGACAAAACCACATGCAG | |||
| d5 | rs65087257 | chr16 | rs65087257-up | CCTTCCTTTTCCACTGTCCA |
| rs65087257-low | CTTAGGGCCTAGGGGCTTTA | |||
| d6 | rs106021355 | chr17 | rs106021355-up | GGTGGTTTATGGCCAGGATA |
| rs106021355-low | AACTGGAGAGGAAAAGGGACA | |||
| d7 | rs106171441 | chr18 | rs106171441-up | TCAATTCTTGGCAGCATCAG |
| rs106171441-low | CTGGCCACTTCTCTGAGTCC | |||
| d8 | rs106777713 | chr19 | rs106777713-up | GTGAAGCGACATCGTCAAGA |
| rs106777713-low | CTTGGCTGTGAGATCGAGGT | |||
| d9 | rs104937988 | chr20 | rs104937988-up | GCTGCACAGAACTGATTCCA |
| rs104937988-low | TGCAAGAATTAAATAAAATCCAAA | |||
| d10 | rs105294203 | chrX | rs105294203-up | ACTCCCCAGAGGATTGTCG |
| rs105294203-low | ATGTTCAAGTGCAACGATGG |
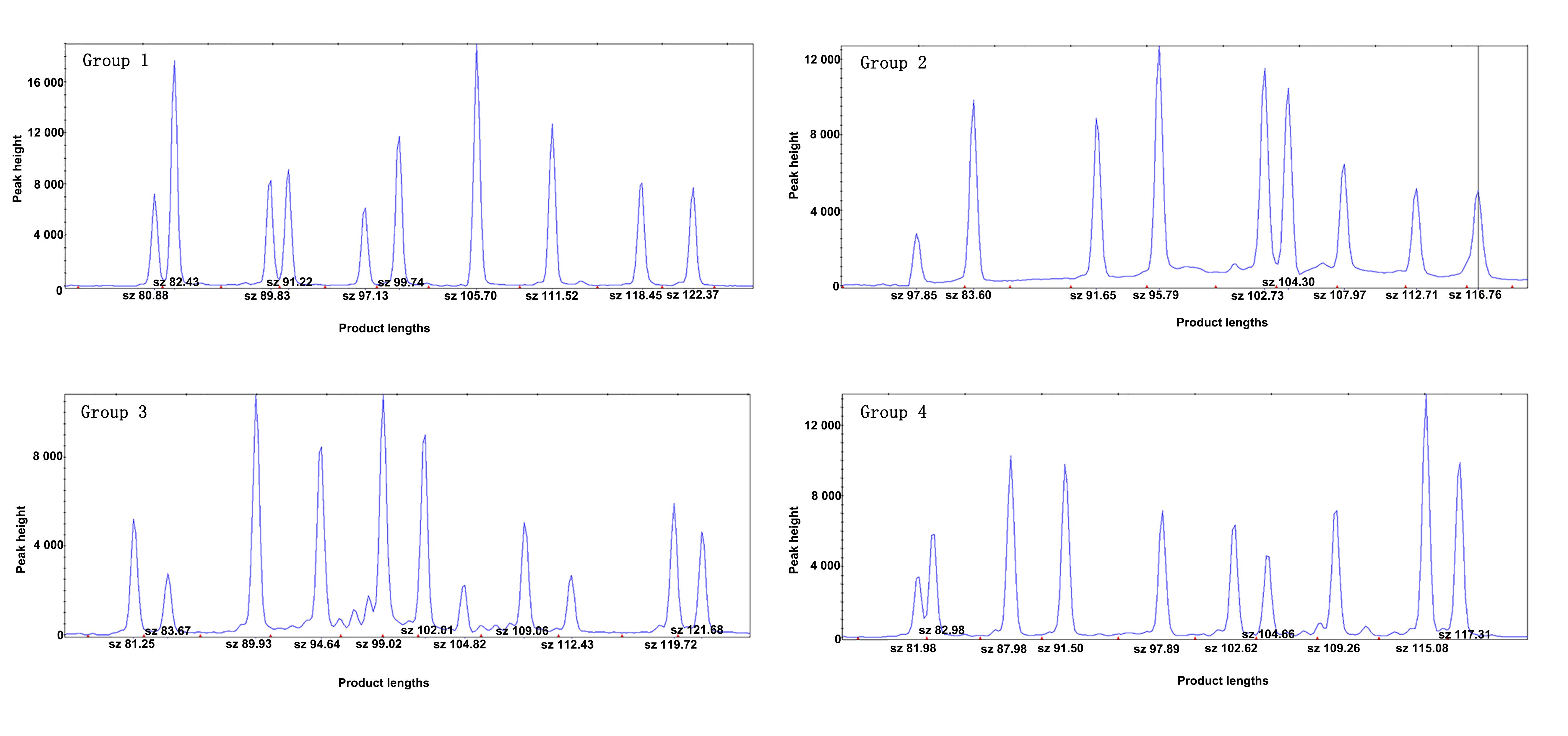
Figure 1 Results of SNP detection protocol (capillary electrophoresis maps of 40 SNP sites for detection, using the DNA of Lewis/BKl rats as template)
大鼠品系 Rat strains | 碱基有差异的SNP位点编号(右上三角)及数量(左下三角) Differences in SNP loci (right-upper triangle) and numbers (left-lower triangle) | ||||
|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | |
| F344/BKl | - | a1/a2/a3/a4/a5/a9/a10/b2/b5/b7/c2/c3/c6/c8/c10/c11/d2/d6/d7/d8/d9 | a1/a2/a4/a7/a9/b4/b8/c1/c2/c3/c6/c9/c11/d2/d7/d8/d10 | a4/b1/b3/b4/b5/b6/b7/c1/c6/c8/c9/d1/d2 | a2/a4/a7/b2/b7/c1/c2/c4/c5/c7/c8/c11/d1/d2/d7/d8 |
| BN/BKl | 21 | - | a3/a5/a7/a10/b2/b4/b5/b7/b8/c1/c8/c9/c10/d6/d9/d10 | a1/a2/a3/a5/a9/a10/b1/b2/b3/b4/b6/c1/c2/c3/c9/c10/c11/d1/d6/d7/d8/d9 | a1/a3/a5/a7/a9/a10/b5/c1/c3/c4/c5/c6/c7/c10/d1/d6/d9 |
| DA/BKl | 17 | 16 | - | a1/a2/a7/a9/b1/b3/b5/b6/b7/b8/c2/c3/c8/c11/d1/d7/d8/d10 | a1/a9/b2/b4/b7/b8/c3/c4/c5/c6/c7/c8/c9/d1/d10 |
| PVG/BKl | 13 | 22 | 18 | - | a2/a7/b1/b2/b3/b4/b5/b6/c2/c4/c5/c6/c7/c9/c11/d7/d8 |
| Lewis/BKl | 16 | 17 | 15 | 17 | - |
Table 2 The number and quantity of the SNP loci with different bases among five rat strains from Lab A
大鼠品系 Rat strains | 碱基有差异的SNP位点编号(右上三角)及数量(左下三角) Differences in SNP loci (right-upper triangle) and numbers (left-lower triangle) | ||||
|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | |
| F344/BKl | - | a1/a2/a3/a4/a5/a9/a10/b2/b5/b7/c2/c3/c6/c8/c10/c11/d2/d6/d7/d8/d9 | a1/a2/a4/a7/a9/b4/b8/c1/c2/c3/c6/c9/c11/d2/d7/d8/d10 | a4/b1/b3/b4/b5/b6/b7/c1/c6/c8/c9/d1/d2 | a2/a4/a7/b2/b7/c1/c2/c4/c5/c7/c8/c11/d1/d2/d7/d8 |
| BN/BKl | 21 | - | a3/a5/a7/a10/b2/b4/b5/b7/b8/c1/c8/c9/c10/d6/d9/d10 | a1/a2/a3/a5/a9/a10/b1/b2/b3/b4/b6/c1/c2/c3/c9/c10/c11/d1/d6/d7/d8/d9 | a1/a3/a5/a7/a9/a10/b5/c1/c3/c4/c5/c6/c7/c10/d1/d6/d9 |
| DA/BKl | 17 | 16 | - | a1/a2/a7/a9/b1/b3/b5/b6/b7/b8/c2/c3/c8/c11/d1/d7/d8/d10 | a1/a9/b2/b4/b7/b8/c3/c4/c5/c6/c7/c8/c9/d1/d10 |
| PVG/BKl | 13 | 22 | 18 | - | a2/a7/b1/b2/b3/b4/b5/b6/c2/c4/c5/c6/c7/c9/c11/d7/d8 |
| Lewis/BKl | 16 | 17 | 15 | 17 | - |
序号 No. | SNP标记名称 Reference SNP ID | 染色体 Chromosome | 不同品系大鼠的基因型 Genotype of different strains of rats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | SHR/Slac | GK/Slac | F344/JclSlac | Wistar/Slac | WKY/Slac | CXB | CHN1 | CHN2 | CHN5 | |||
| a1 | rs65970293 | chr1 | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | T/T | T/T | T/T | T/T | T/T |
| a2 | rs106996961 | chr2 | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | T/T |
| a3 | rs63860341 | chr3 | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| a4 | rs64750237 | chr4 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| a5 | rs107261116 | chr5 | C/C | G/G | C/C | C/C | C/C | G/G | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G |
| a6 | rs64102787 | chr6 | C/C | C/C | C/C | C/C | C/C | A/A | A/A | C/C | C/C | A/A | C/C | C/C | C/C | A/A |
| a7 | rs105187832 | chr7 | G/G | G/G | T/T | G/G | T/T | G/G | T/T | G/G | T/T | T/T | G/G | G/G | G/G | G/G |
| a8 | rs64682441 | chr9 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| a9 | rs65067166 | chr10 | C/C | G/G | G/G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G | G/G | G/G |
| a10 | rs8167985 | chr11 | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| b1 | rs106672258 | chr12 | T/T | T/T | T/T | C/C | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| b2 | rs65921305 | chr13 | G/G | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | T/T |
| b3 | rs106458396 | chr15 | T/T | T/T | T/T | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| b4 | rs106888236 | chr16 | C/C | C/C | A/A | A/A | C/C | A/A | A/A | C/C | A/A | C/C | C/C | A/A | A/A | C/C |
| b5 | rs63814610 | chr17 | T/T | C/C | T/T | C/C | T/T | T/T | T/T | T/T | C/C | T/T | C | T/T | T/T | T/T |
| b6 | rs8152541 | chr18 | G/G | G/G | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | A/A | A/A | G/G | A/A |
| b7 | rs105824121 | chr19 | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | G/G | G/G | A/A | A/A | A/A | G/G |
| b8 | rs13453252 | chr20 | G/G | G/G | C/C | G/G | G/G | G/G | C/C | G/G | C/C | C/C | G/G | G/G | G/G | G/G |
| b9 | rs65108991 | chrX | A/A | A/A | A/A | A/A | A/A | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A |
| c1 | rs106891788 | chr1 | C/C | C/C | T/T | T/T | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| c2 | rs107493604 | chr2 | A/A | G/G | G/G | A/A | G/G | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | G/G |
| c3 | rs65632519 | chr3 | T/T | A/A | A/A | T/T | T/T | A/A | A/A | T/T | T/T | T/T | A | T/T | T/T | T/T |
| c4 | rs17163178 | chr4 | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | T/T | T/T | C/C |
| c5 | rs105170847 | chr5 | C/C | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| c6 | rs106547562 | chr6 | A/A | C/C | C/C | C/C | A/A | C/C | C/C | A/A | A/A | C/C | A/A | A/A | A/A | C/C |
| c7 | rs65502317 | chr7 | C/C | C/C | C/C | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A | A/A | C/C |
| c8 | rs105694545 | chr8 | G/G | A/A | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | G/G | A/A | A/A |
| c9 | rs65801920 | chr9 | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| c10 | rs105168947 | chr10 | C/C | T/T | C/C | C/C | C/C | T/T | T/T | C/C | C/C | C/C | T/T | T/T | T/T | C/C |
| c11 | rs106880606 | chr11 | G/G | T/T | T/T | G/G | T/T | T/T | T/T | G/G | T/T | T/T | T/T | T/T | T/T | G/G |
| d1 | rs8158863 | chr12 | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | A/A | G/G | A/A | A/A | A/A | G/G |
| d2 | rs105965886 | chr13 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C |
| d3 | rs63919332 | chr14 | C/C | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | C/C |
| d4 | rs65809306 | chr15 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | T/T | T/T | T/T | C/C |
| d5 | rs65087257 | chr16 | A/A* | A/A | A/A | A/A | A/A | A/A | C/C | C/C* | C/C | A/A | C/C | A/A | A/A | C/C |
| d6 | rs106021355 | chr17 | T/T | C/C | T/T | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| d7 | rs106171441 | chr18 | G/G | A/A | A/A | G/G | A/A | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A | A/A |
| d8 | rs106777713 | chr19 | G/G* | A/A | A/A | G/G | A/A | T/T | C/C | T/T* | T/T | C/C | C/C | T/T | T/T | T/T |
| d9 | rs104937988 | chr20 | T/T | G/G | T/T | T/T | T/T | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | G/G |
| d10 | rs105294203 | chrX | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | G/G | G/G | G/G |
Table 3 Multiplex PCR-ligase detection reaction (LDR) test results of inbred rat strains from different domestic animal
序号 No. | SNP标记名称 Reference SNP ID | 染色体 Chromosome | 不同品系大鼠的基因型 Genotype of different strains of rats | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F344/BKl | BN/BKl | DA/BKl | PVG/BKl | Lewis/BKl | SHR/Slac | GK/Slac | F344/JclSlac | Wistar/Slac | WKY/Slac | CXB | CHN1 | CHN2 | CHN5 | |||
| a1 | rs65970293 | chr1 | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | T/T | T/T | T/T | T/T | T/T |
| a2 | rs106996961 | chr2 | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | T/T |
| a3 | rs63860341 | chr3 | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| a4 | rs64750237 | chr4 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| a5 | rs107261116 | chr5 | C/C | G/G | C/C | C/C | C/C | G/G | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G |
| a6 | rs64102787 | chr6 | C/C | C/C | C/C | C/C | C/C | A/A | A/A | C/C | C/C | A/A | C/C | C/C | C/C | A/A |
| a7 | rs105187832 | chr7 | G/G | G/G | T/T | G/G | T/T | G/G | T/T | G/G | T/T | T/T | G/G | G/G | G/G | G/G |
| a8 | rs64682441 | chr9 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| a9 | rs65067166 | chr10 | C/C | G/G | G/G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/G | G/G | G/G |
| a10 | rs8167985 | chr11 | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| b1 | rs106672258 | chr12 | T/T | T/T | T/T | C/C | T/T | C/C | C/C | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| b2 | rs65921305 | chr13 | G/G | T/T | G/G | G/G | T/T | G/G | G/G | G/G | T/T | G/G | G/G | G/G | G/G | T/T |
| b3 | rs106458396 | chr15 | T/T | T/T | T/T | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| b4 | rs106888236 | chr16 | C/C | C/C | A/A | A/A | C/C | A/A | A/A | C/C | A/A | C/C | C/C | A/A | A/A | C/C |
| b5 | rs63814610 | chr17 | T/T | C/C | T/T | C/C | T/T | T/T | T/T | T/T | C/C | T/T | C | T/T | T/T | T/T |
| b6 | rs8152541 | chr18 | G/G | G/G | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | A/A | A/A | G/G | A/A |
| b7 | rs105824121 | chr19 | G/G | A/A | G/G | A/A | A/A | G/G | G/G | G/G | G/G | G/G | A/A | A/A | A/A | G/G |
| b8 | rs13453252 | chr20 | G/G | G/G | C/C | G/G | G/G | G/G | C/C | G/G | C/C | C/C | G/G | G/G | G/G | G/G |
| b9 | rs65108991 | chrX | A/A | A/A | A/A | A/A | A/A | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A |
| c1 | rs106891788 | chr1 | C/C | C/C | T/T | T/T | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | T/T | T/T |
| c2 | rs107493604 | chr2 | A/A | G/G | G/G | A/A | G/G | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | G/G |
| c3 | rs65632519 | chr3 | T/T | A/A | A/A | T/T | T/T | A/A | A/A | T/T | T/T | T/T | A | T/T | T/T | T/T |
| c4 | rs17163178 | chr4 | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | T/T | T/T | C/C |
| c5 | rs105170847 | chr5 | C/C | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| c6 | rs106547562 | chr6 | A/A | C/C | C/C | C/C | A/A | C/C | C/C | A/A | A/A | C/C | A/A | A/A | A/A | C/C |
| c7 | rs65502317 | chr7 | C/C | C/C | C/C | C/C | A/A | A/A | A/A | C/C | A/A | A/A | A/A | A/A | A/A | C/C |
| c8 | rs105694545 | chr8 | G/G | A/A | G/G | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | G/G | A/A | A/A |
| c9 | rs65801920 | chr9 | T/T | T/T | C/C | C/C | T/T | C/C | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T |
| c10 | rs105168947 | chr10 | C/C | T/T | C/C | C/C | C/C | T/T | T/T | C/C | C/C | C/C | T/T | T/T | T/T | C/C |
| c11 | rs106880606 | chr11 | G/G | T/T | T/T | G/G | T/T | T/T | T/T | G/G | T/T | T/T | T/T | T/T | T/T | G/G |
| d1 | rs8158863 | chr12 | A/A | A/A | A/A | G/G | G/G | G/G | G/G | A/A | A/A | G/G | A/A | A/A | A/A | G/G |
| d2 | rs105965886 | chr13 | C/C | T/T | T/T | T/T | T/T | C/C | C/C | C/C | T/T | C/C | C/C | C/C | C/C | C/C |
| d3 | rs63919332 | chr14 | C/C | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/T | C/C | C/C | C/C | C/C |
| d4 | rs65809306 | chr15 | T/T | T/T | T/T | T/T | T/T | C/C | C/C | T/T | T/T | C/C | T/T | T/T | T/T | C/C |
| d5 | rs65087257 | chr16 | A/A* | A/A | A/A | A/A | A/A | A/A | C/C | C/C* | C/C | A/A | C/C | A/A | A/A | C/C |
| d6 | rs106021355 | chr17 | T/T | C/C | T/T | T/T | T/T | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T |
| d7 | rs106171441 | chr18 | G/G | A/A | A/A | G/G | A/A | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A | A/A |
| d8 | rs106777713 | chr19 | G/G* | A/A | A/A | G/G | A/A | T/T | C/C | T/T* | T/T | C/C | C/C | T/T | T/T | T/T |
| d9 | rs104937988 | chr20 | T/T | G/G | T/T | T/T | T/T | G/G | G/G | T/T | T/T | G/G | G/G | G/G | G/G | G/G |
| d10 | rs105294203 | chrX | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | A/A | G/G | G/G | G/G | G/G | G/G |
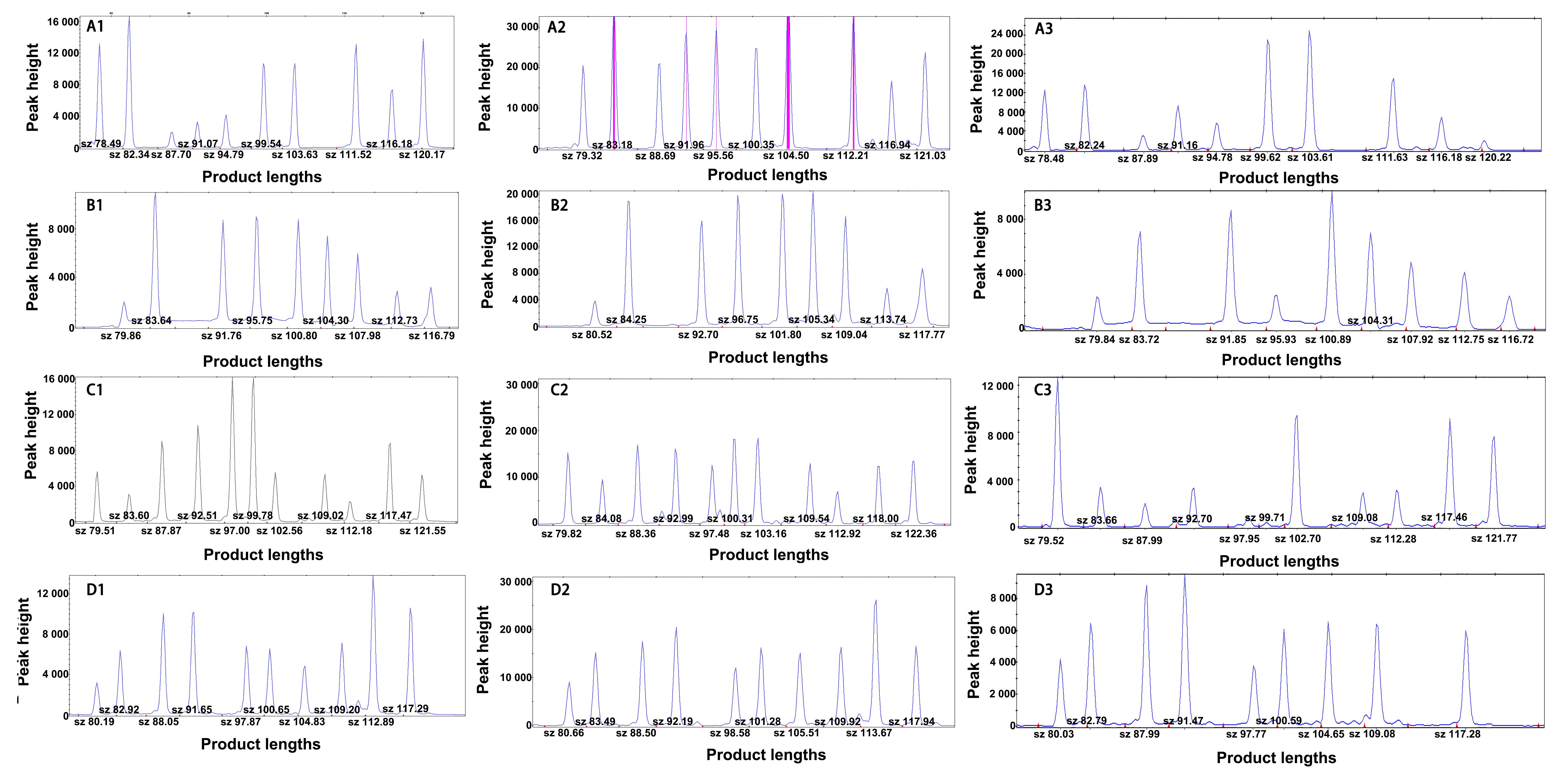
Figure 2 Application results of three DNA polymerases in the multiple PCR-LDR detection scheme of rat SNP (using BN/BKl rats as an example)Note: A1-A3, Results of the first group of SNP loci detected using enzyme 1, enzyme 2, and enzyme 3, respectively; B1-B3, Results of the second group of SNP loci detected for enzyme 1, enzyme 2, and enzyme 3; C1-C3, Results of the third group of SNP loci detected by enzyme 1, enzyme 2, and enzyme 3; D1-D3, Results of the fourth group of SNP loci detected for enzyme 1, enzyme 2, and enzyme 3.
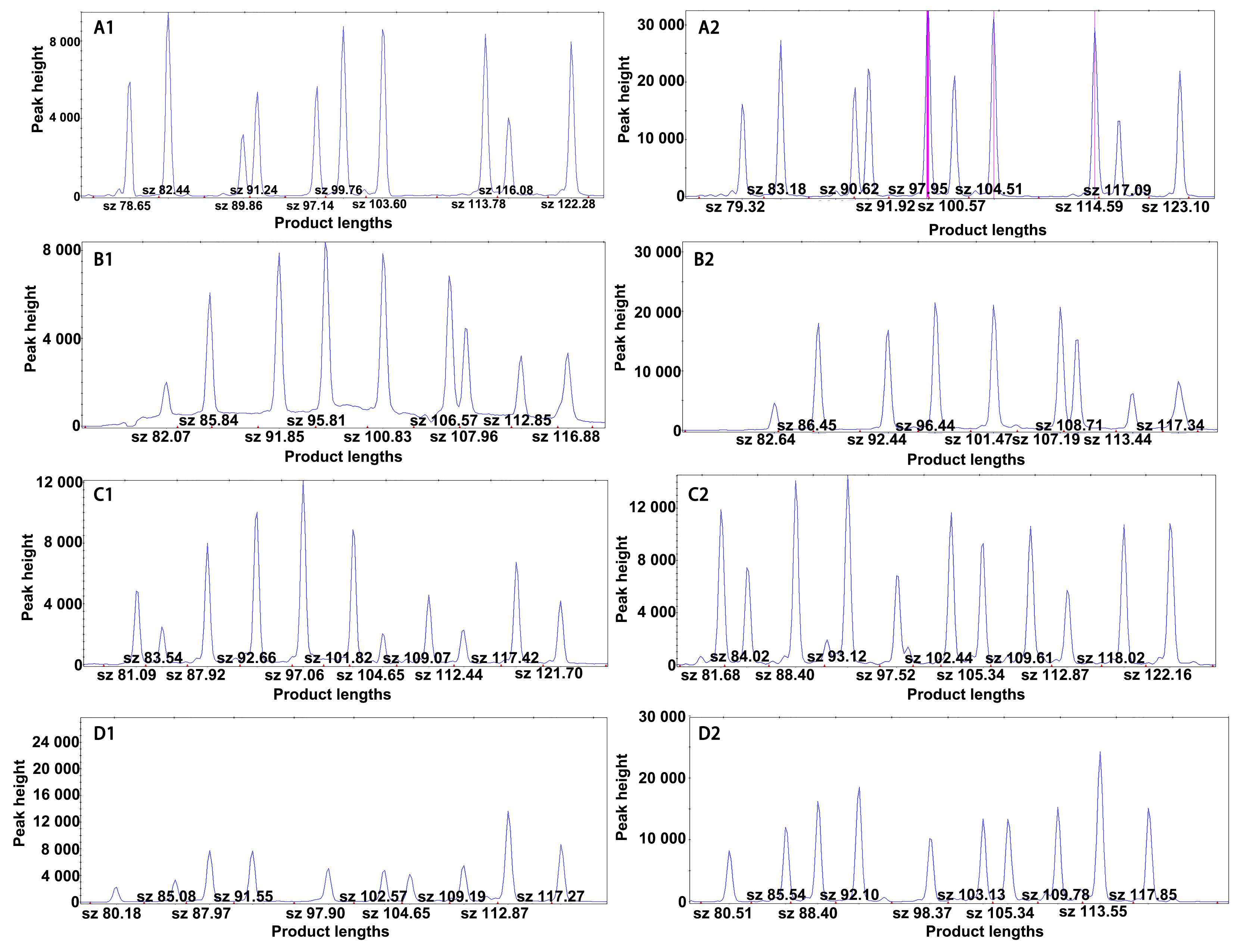
Figure 3 Comparison of multiplex PCR-ligase detection reaction (LDR) results among different laboratories (using CXB rats as an example)Note: A1-D1, electrophoretic peak maps of SNP sites in group 1- 4 tested by our institution. A2-D2, electrophoretic peak maps of SNP sites in group 1- 4 tested by the third-party laboratory.
| 1 | BENAVIDES F, RÜLICKE T, PRINS J B, et al. Genetic quality assurance and genetic monitoring of laboratory mice and rats: FELASA Working Group Report[J]. Lab Anim, 2020, 54(2):135-148. DOI: 10.1177/0023677219867719 . |
| 2 | HOFFMAN H A. 实验动物遗传质量的控制重点在遗传监测[J]. 上海实验动物科学, 1987, 7(1): 55-57. |
| HOFFMAN H A. The key to controlling the genetic quality of experimental animals is genetic monitoring[J]. Shanghai Lab Anim Sci, 1987, 7(1): 55-57. | |
| 3 | 国家市场监督管理总局, 国家标准化管理委员会. 实验动物 遗传质量控制: GB 14923—2022[S]. 北京: 中国标准出版社, 2022. |
| State Administration for Market Regulation, Standardization Administration of the People's Republic of China. Laboratory animal-Genetic quality control: GB 14923-2022[S]. Beijing: Standards Press of China, 2022 | |
| 4 | 杨玲焰,王立鹏,时长军. 一种基于KASP的大鼠遗传质量监控SNP标记分型方法及试剂盒: CN201910648812X[P/OL]. 2019-11-15[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| YANG L Y, WANG L P, SHI C J. A SNP marker typing method and kit based KASP for monitoring rat genetic quality: CN201910648812X[P/OL]. 2019-11-15[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 5 | 琚存祥, 赵静, 马秀英, 等. 一种近交系遗传质量监控的SNP快速检测方法和SNP位点及其引物: CN108588236A[P/OL]. 2018-09-28[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| JU C X, ZHAO J, MA X Y, et al. Rapid SNP detection method for genetic quality monitoring of inbred lines and SNP sites and primers: CN108588236A[P/OL]. 2018-09-28[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 6 | 琚存祥, 赵静, 杨旭乐, 等. 一组用于BALB/cJ近交系小鼠遗传质量监控的SNP位点及其引物组合和应用: CN110358847A[P/OL]. 2019-10-22[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| JU C X, ZHAO J, YANG X L, et al. A set of SNP sites and primer combinations and applications for genetic quality control in BALB/cJ inbred mice: CN110358847A[P/OL]. 2019-10-22[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 7 | 赵静, 琚存祥, 马秀英, 等. 一组用于CBA/CaJ近交系小鼠遗传质量监控的SNP位点及其引物组合和应用: CN109609659A[P/OL]. 2019-04-12[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. |
| ZHAO J, JU C X, MA X Y, et al. A set of SNP sites and their primer combinations and applications for genetic quality monitoring in CBA/CaJ inbred mice: CN109609659A[P/OL]. 2019-04-12[2023-05-10]. http://epub.cnipa.gov.cn/Dxb/IndexQuery. | |
| 8 | 王刚, 李凯, 周宇荀, 等. 新型通用探针LDR分型技术的开发及细胞色素P450基因多位点分型[J]. 华东理工大学学报(自然科学版), 2008, 34(4):503-508. DOI: 10.3969/j.issn.1006-3080.2006.09.008 . |
| WANG G, LI K, ZHOU Y X, et al. Improvement of novel LDR genotyping system based on universal probe and the SNPs typing of human P450[J]. J East China Univ Sci Technol Nat Sci Ed, 2008, 34(4):503-508. DOI: 10.3969/j.issn.1006-3080.2006.09.008 . | |
| 9 | 徐园, 钱强, 鲍世民, 等. 基于高通量靶向测序的大鼠遗传质量检测方案建立[J]. 生物化工, 2020, 6(4): 65-69. DOI: 10.3969/j.issn.2096-0387.2020.04.016 . |
| XU Y, QIAN Q, BAO S M, et al. Establishment of rat genetic quality detection scheme based on high throughput targeted sequencing[J]. Biol Chem Eng, 2020, 6(4): 65-69. DOI: 10.3969/j.issn.2096-0387.2020.04.016 . | |
| 10 | SAAR K, BECK A, BIHOREAU M T, et al. SNP and haplotype mapping for genetic analysis in the rat[J]. Nat Genet, 2008, 40(5):560-566. DOI: 10.1038/ng.124 . |
| 11 | PETKOV P M, DING Y M, CASSELL M A, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships[J]. Genome Res, 2004, 14(9):1806-1811. DOI: 10.1101/gr.2825804 . |
| 12 | TAFT R A, DAVISSON M, WILES M V. Know thy mouse[J]. Trends Genet, 2006, 22(12):649-653. DOI: 10.1016/j.tig.2006.09.010 . |
| 13 | MEKADA K, HIROSE M, MURAKAMI A, et al. Development of SNP markers for C57BL/6N-derived mouse inbred strains[J]. Exp Anim, 2015, 64(1):91-100. DOI: 10.1538/expanim.14-0061 . |
| [1] | QIAN Qiang, XU Yuan, WANG Ya-heng, ZHOU Yu-xun, XIAO Jun-hua, HAN Ling, BAO Shi-ming, LI Kai. Mouse Genetic Quality Monitoring Method Establishment Based on Next-generation Sequencing through Multiplex PCR [J]. Laboratory Animal and Comparative Medicine, 2019, 39(2): 111-117. |
| [2] | ZHAO Li-ya, ZHANG Rong, ZHAO Ying, XING Zheng-hong, CHEN Guo-qiang. Application of Single Nucleotide Polymorphism Genotyping Panel in Genetic Monitoring of Laboratory Mice [J]. Laboratory Animal and Comparative Medicine, 2016, 36(1): 24-31. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||