
Laboratory Animal and Comparative Medicine ›› 2025, Vol. 45 ›› Issue (6): 773-783.DOI: 10.12300/j.issn.1674-5817.2025.138
• Invertebrate Laboratory Animals: Mosquito • Previous Articles Next Articles
YUN Jiaqi1,2, MA Qin1,2, WANG Guandong1, SUN Peilu1, WANG Yiguan1, WANG Sibao1( )(
)( )
)
Received:2025-09-05
Revised:2025-10-14
Online:2025-12-25
Published:2025-12-19
Contact:
WANG Sibao
CLC Number:
YUN Jiaqi,MA Qin,WANG Guandong,et al. Research Advances and Challenges of Gene Drive Technology in Mosquito-Borne Disease Control[J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 773-783. DOI: 10.12300/j.issn.1674-5817.2025.138.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2025.138
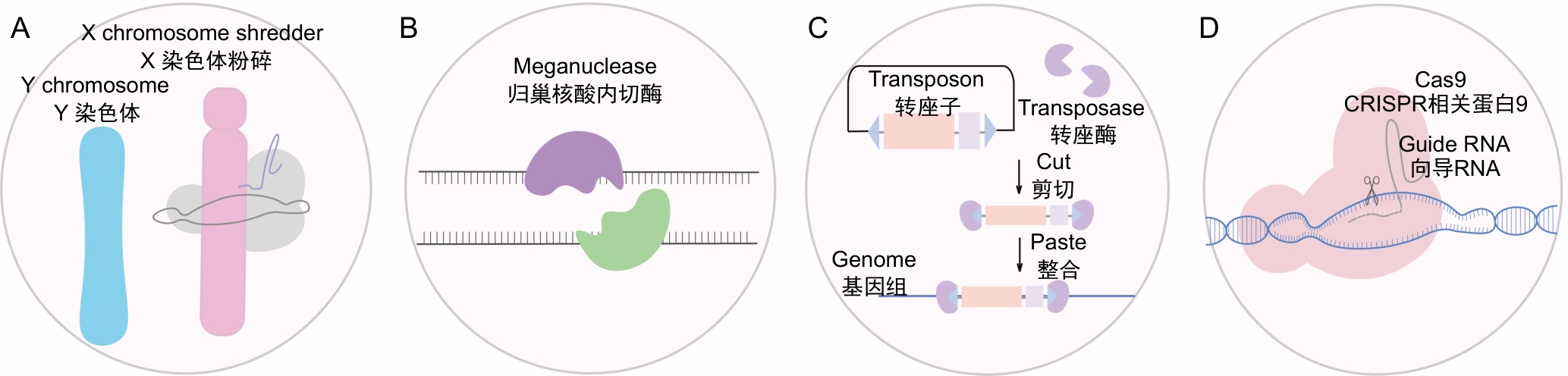
Figure 1 Gene drive types based on different driver elementsNote: A, Meiosis-mediated gene drive, X chromosome shattering leads to a strong sex ratio distortion, with offspring tending to be male; B, Gene drive based on meganuclease, meganuclease recognizes and cuts specific DNA sequence, triggering the homologous recombination repair mechanism; C, Gene drive based on transposon and transposase systems, transposases catalyze the insertion and movement of transposons, randomly inserting target genes into the host genome; D, Gene drive based on the CRISPR/Cas9 system, the drive gene is precisely inserted into the target gene locus, and the Cas9 protein continuously cuts the wild-type allele.
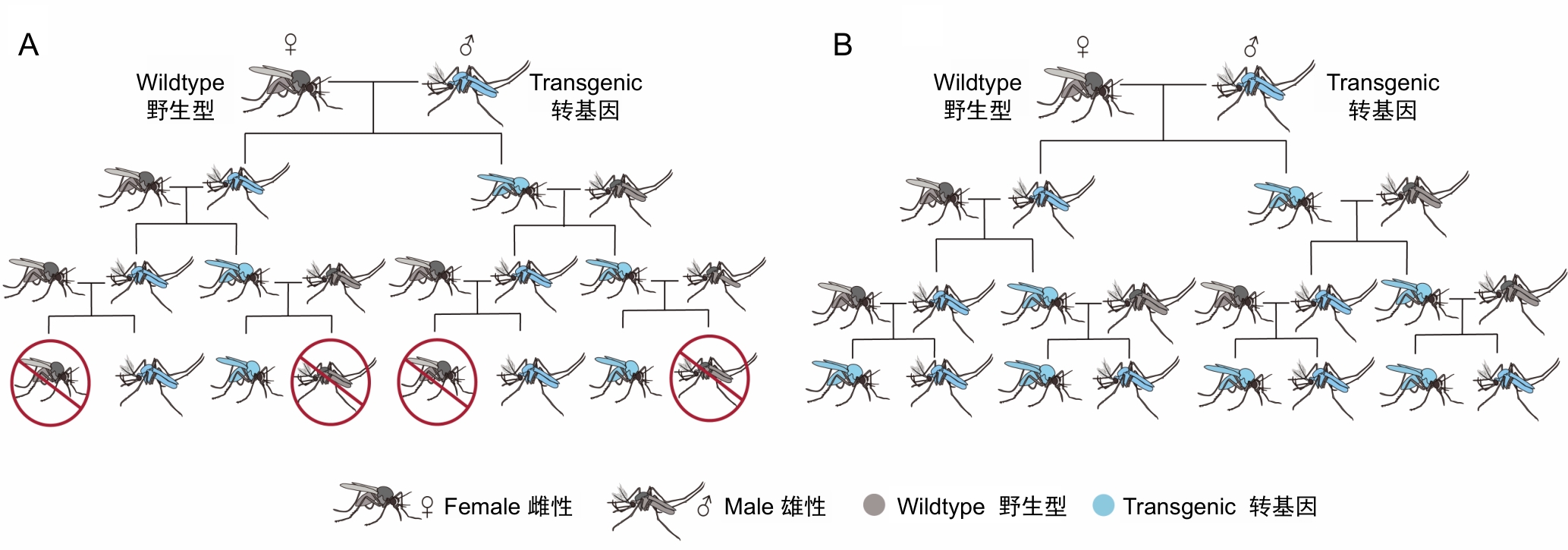
Figure 2 Gene drive types based on functional classificationNote: A, Population suppression gene drive, by designing genetic elements with "selfish inheritance" characteristics, rapidly spreads lethal or reproduction-interfering genes in the mosquito population, ultimately achieving population suppression; B, Population replacement gene drive: through gene editing, endows mosquitoes with disease-resistance characteristics, blocking the transmission chain of pathogens without significantly altering the population size.
| 减数分裂介导的基因驱动 | Galizi R, | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Kyrou K, | ||
| Hammond A, et al. (2016)[ | |||
| fibrinogen-related protein 1 | Dong Y, et al. (2018)[ | ||
| cardinal | Carballar-lejarazu R, et al. (2020)[ | ||
| single chain variable fragment | Carballar-lejarazu R, et al. (2023)[ | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Xu X, et al. (2025)[ | ||
| Gantz V M, et al. (2015)[ | |||
| CRISPR/Cas9介导的基因驱动 | Green E I, et al. (2023)[ | ||
致倦库蚊 Culex quinquefasciatus | CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | Feng X C, et al. (2021)[ | |
| CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | O'Leary S, et al. (2020)[ | ||
| Li M, et al. (2020)[ | |||
| 归巢核酸内切酶介导的基因驱动 | Franz A W, et al. (2006)[ Williams A E, et al. (2020)[ | ||
| 转座子介导的基因驱动 | Buchman A, et al. (2020)[ |
Table 1 Main advances of gene drive technology in mosquito-borne disease control
| 减数分裂介导的基因驱动 | Galizi R, | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Kyrou K, | ||
| Hammond A, et al. (2016)[ | |||
| fibrinogen-related protein 1 | Dong Y, et al. (2018)[ | ||
| cardinal | Carballar-lejarazu R, et al. (2020)[ | ||
| single chain variable fragment | Carballar-lejarazu R, et al. (2023)[ | ||
CRISPR/Cas9介导的基因驱动 CRISPR/Cas9介导的基因驱动 | Xu X, et al. (2025)[ | ||
| Gantz V M, et al. (2015)[ | |||
| CRISPR/Cas9介导的基因驱动 | Green E I, et al. (2023)[ | ||
致倦库蚊 Culex quinquefasciatus | CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | Feng X C, et al. (2021)[ | |
| CRISPR/Cas9介导的基因驱动CRISPR/Cas9介导的基因驱动 | O'Leary S, et al. (2020)[ | ||
| Li M, et al. (2020)[ | |||
| 归巢核酸内切酶介导的基因驱动 | Franz A W, et al. (2006)[ Williams A E, et al. (2020)[ | ||
| 转座子介导的基因驱动 | Buchman A, et al. (2020)[ |
| [1] | World Health Organization. Vector-borne diseases[Z/OL]. (2024-09-26)[2025-08-21] . |
| [2] | World Health Organization. Dengue[Z/OL]. (2025-08-21)[2025-08-21]. . |
| [3] | World Health Organization. New WHO guidelines for clinical management of arboviral diseases: dengue, chikungunya, Zika and yellow fever[Z/OL]. (2025-07-10)[2025-08-21]. . |
| [4] | KITTAYAPONG P, NINPHANOMCHAI S, LIMOHPASMANEE W, et al. Combined sterile insect technique and incompatible insect technique: the first proof-of-concept to suppress Aedes aegypti vector populations in semi-rural settings in Thailand[J]. PLoS Negl Trop Dis, 2019, 13(10):e0007771. DOI:10.1371/journal.pntd.0007771 . |
| [5] | HARRIS A F, MCKEMEY A R, NIMMO D, et al. Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes[J]. Nat Biotechnol, 2012, 30(9):828-830. DOI:10.1038/nbt.2350 . |
| [6] | MCMENIMAN C J, LANE R V, CASS B N, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti [J]. Science, 2009, 323(5910):141-144. DOI:10.1126/science.1165326 . |
| [7] | PHUC H K, ANDREASEN M H, BURTON R S, et al. Late-acting dominant lethal genetic systems and mosquito control[J]. BMC Biol, 2007, 5:11. DOI:10.1186/1741-7007-5-11 . |
| [8] | HOFFMANN A A, MONTGOMERY B L, POPOVICI J, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission[J]. Nature, 2011, 476(7361):454-457. DOI:10.1038/nature10356 . |
| [9] | AMUZU H E, TSYGANOV K, KOH C, et al. Wolbachia enhances insect-specific flavivirus infection in Aedes aegypti mosquitoes[J]. Ecol Evol, 2018, 8(11):5441-5454. DOI:10.1002/ece3.4066 . |
| [10] | ZÉLÉ F, NICOT A, BERTHOMIEU A, et al. Wolbachia increases susceptibility to Plasmodium infection in a natural system[J]. Proc Biol Sci, 2014, 281(1779):20132837. DOI:10.1098/rspb.2013.2837 . |
| [11] | BURT A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations[J]. Proc Biol Sci, 2003, 270(1518):921-928. DOI:10.1098/rspb.2002.2319 . |
| [12] | AKBARI O S, BELLEN H J, BIER E, et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory[J]. Science, 2015, 349(6251):927-929. DOI:10.1126/science.aac7932 . |
| [13] | MARSHALL J M, HAY B A. Confinement of gene drive systems to local populations: a comparative analysis[J]. J Theor Biol, 2012, 294:153-171. DOI:10.1016/j.jtbi.2011.10.032 . |
| [14] | CHAMPER J, BUCHMAN A, AKBARI O S. Cheating evolution: engineering gene drives to manipulate the fate of wild populations[J]. Nat Rev Genet, 2016, 17(3):146-159. DOI:10.1038/nrg.2015.34 . |
| [15] | JAMES A A. Gene drive systems in mosquitoes: rules of the road[J]. Trends Parasitol, 2005, 21(2):64-67. DOI:10.1016/j.pt.2004.11.004 . |
| [16] | O'BROCHTA D A, ALFORD R T, PILITT K L, et al. piggyBac transposon remobilization and enhancer detection in Anopheles mosquitoes[J]. Proc Natl Acad Sci USA, 2011, 108(39):16339-16344. DOI:10.1073/pnas.1110628108 . |
| [17] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121):819-823. DOI:10.1126/science.1231143 . |
| [18] | KYROU K, HAMMOND A M, GALIZI R, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes[J]. Nat Biotechnol, 2018, 36(11):1062-1066. DOI:10.1038/nbt.4245 . |
| [19] | HAMMOND A, GALIZI R, KYROU K, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae [J]. Nat Biotechnol, 2016, 34(1):78-83. DOI:10.1038/nbt.3439 . |
| [20] | DONG Y M, SIMÕES M L, MAROIS E, et al. CRISPR/Cas9-mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection[J]. PLoS Pathog, 2018, 14(3):e1006898. DOI:10.1371/journal.ppat.1006898 . |
| [21] | CARBALLAR-LEJARAZÚ R, OGAUGWU C, TUSHAR T, et al. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae [J]. Proc Natl Acad Sci USA, 2020, 117(37):22805-22814. DOI:10.1073/pnas.2010214117 . |
| [22] | GALIZI R, DOYLE L A, MENICHELLI M, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito[J]. Nat Commun, 2014, 5:3977. DOI:10.1038/ncomms4977 . |
| [23] | XU X J, CHEN J H, WANG Y, et al. Gene drive-based population suppression in the malaria vector Anopheles stephensi [J]. Nat Commun, 2025, 16(1):1007. DOI:10.1038/s41467-025-56290-2 . |
| [24] | LI Z Q, DONG Y M, YOU L, et al. Driving a protective allele of the mosquito FREP1 gene to combat malaria[J]. Nature, 2025, 645(8081):746-754. DOI:10.1038/s41586-025-09283-6 . |
| [25] | GREEN E I, JAOUEN E, KLUG D, et al. A population modification gene drive targeting both Saglin and Lipophorin impairs Plasmodium transmission in Anopheles mosquitoes[J]. eLife, 2023, 12:e93142. DOI:10.7554/eLife.93142 . |
| [26] | CARBALLAR-LEJARAZÚ R, DONG Y M, PHAM T B, et al. Dual effector population modification gene-drive strains of the African malaria mosquitoes, Anopheles gambiae and Anopheles coluzzii [J]. Proc Natl Acad Sci USA, 2023, 120(29):e2221118120. DOI:10.1073/pnas.2221118120 . |
| [27] | GANTZ V M, JASINSKIENE N, TATARENKOVA O, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi [J]. Proc Natl Acad Sci USA, 2015, 112(49):E6736-E6743. DOI:10.1073/pnas.1521077112 . |
| [28] | MUÑOZ D, JIMENEZ A, MARINOTTI O, et al. The AeAct-4 gene is expressed in the developing flight muscles of female Aedes aegypti [J]. Insect Mol Biol, 2004, 13(5):563-568. DOI:10.1111/j.0962-1075.2004.00519.x . |
| [29] | O'LEARY S, ADELMAN Z N. CRISPR/Cas9 knockout of female-biased genes AeAct-4 or myo-fem in Ae. aegypti results in a flightless phenotype in female, but not male mosquitoes[J]. PLoS Negl Trop Dis, 2020, 14(12):e0008971. DOI:10.1371/journal.pntd.0008971 . |
| [30] | FRANZ A W E, SANCHEZ-VARGAS I, ADELMAN Z N, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti [J]. Proc Natl Acad Sci USA, 2006, 103(11):4198-4203. DOI:10.1073/pnas.0600479103 . |
| [31] | WILLIAMS A E, SANCHEZ-VARGAS I, REID W R, et al. The antiviral small-interfering RNA pathway induces zika virus resistance in transgenic Aedes aegypti [J]. Viruses, 2020, 12(11):1231. DOI:10.3390/v12111231 . |
| [32] | BUCHMAN A, GAMEZ S, LI M, et al. Broad dengue neutralization in mosquitoes expressing an engineered antibody[J]. PLoS Pathog, 2020, 16(1):e1008103. DOI:10.1371/journal.ppat.1008103 . |
| [33] | REID W R, OLSON K E, FRANZ A W E. Current effector and gene-drive developments to engineer arbovirus-resistant Aedes aegypti (Diptera: Culicidae) for a sustainable population replacement strategy in the field[J]. J Med Entomol, 2021, 58(5):1987-1996. DOI:10.1093/jme/tjab030 . |
| [34] | LI M, YANG T, KANDUL N P, et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti [J]. eLife, 2020, 9:e51701. DOI:10.7554/eLife.51701 . |
| [35] | FENG X C, LÓPEZ DEL AMO V, MAMELI E, et al. Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes[J]. Nat Commun, 2021, 12(1):2960. DOI:10.1038/s41467-021-23239-0 . |
| [36] | FENG X C, KAMBIC L, NISHIMOTO J H K, et al. Evaluation of gene knockouts by CRISPR as potential targets for the genetic engineering of the mosquito Culex quinquefasciatus [J]. CRISPR J, 2021, 4(4):595-608. DOI:10.1089/crispr.2021.0028 . |
| [37] | HARVEY-SAMUEL T, FENG X C, OKAMOTO E M, et al. CRISPR-based gene drives generate super-Mendelian inheritance in the disease vector Culex quinquefasciatus [J]. Nat Commun, 2023, 14(1):7561. DOI:10.1038/s41467-023-41834-1 . |
| [38] | KANDUL N P, LIU J R, BUCHMAN A, et al. Assessment of a split homing based gene drive for efficient knockout of multiple genes[J]. G3 (Bethesda), 2020, 10(2):827-837. DOI:10.1534/g3.119.400985 . |
| [39] | CHAMPER J, REEVES R, OH S Y, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations[J]. PLoS Genet, 2017, 13(7):e1006796. DOI:10.1371/journal.pgen.1006796 . |
| [40] | NOBLE C, ADLAM B, CHURCH G M, et al. Current CRISPR gene drive systems are likely to be highly invasive in wild populations[J]. eLife, 2018, 7:e33423. DOI:10.7554/eLife.33423 . |
| [41] | PROWSE T A A, CASSEY P, ROSS J V, et al. Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates[J]. Proc Biol Sci, 2017, 284(1860):20170799. DOI: 10.1098/rspb.2017.0799 . |
| [42] | CHAMPER J, LIU J X, OH S Y, et al. Reducing resistance allele formation in CRISPR gene drive[J]. Proc Natl Acad Sci USA, 2018, 115(21):5522-5527. DOI:10.1073/pnas.1720354115 . |
| [43] | VELLA M R, GUNNING C E, LLOYD A L, et al. Evaluating strategies for reversing CRISPR-Cas9 gene drives[J]. Sci Rep, 2017, 7(1):11038. DOI: 10.1038/s41598-017-10633-2 . |
| [44] | ESVELT K M, SMIDLER A L, CATTERUCCIA F, et al. Concerning RNA-guided gene drives for the alteration of wild populations[J]. eLife, 2014, 3:e03401. DOI: 10.7554/eLife.03401 . |
| [45] | WU B, LUO L Q, GAO X J. Cas9-triggered chain ablation of cas9 as a gene drive brake[J]. Nat Biotechnol, 2016, 34(2):137-138. DOI:10.1038/nbt.3444 . |
| [46] | ADOLFI A, GANTZ V M, JASINSKIENE N, et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi [J]. Nat Commun, 2020, 11(1):5553. DOI: 10.1038/s41467-020-19426-0 . |
| [47] | SÁNCHEZ C H M, WU S L, BENNETT J B, et al. MGDrivE: a modular simulation framework for the spread of gene drives through spatially explicit mosquito populations[J]. Meth Ecol Evol, 2020, 11(2):229-239. DOI:10.1111/2041-210X.13318 . |
| [48] | NORTH A R, BURT A, GODFRAY H C J. Modelling the suppression of a malaria vector using a CRISPR-Cas9 gene drive to reduce female fertility[J]. BMC Biol, 2020, 18(1):98. DOI: 10.1186/s12915-020-00834-z . |
| [49] | OBERHOFER G, IVY T, HAY B A. Gene drive that results in addiction to a temperature-sensitive version of an essential gene triggers population collapse in Drosophila [J]. Proc Natl Acad Sci USA, 2021, 118(49):e2107413118. DOI: 10.1073/pnas.2107413118 . |
| [50] | WANG G D, VEGA-RODRÍGUEZ J, DIABATE A, et al. Clock genes and environmental cues coordinate Anopheles pheromone synthesis, swarming, and mating[J]. Science, 2021, 371(6527):411-415. DOI:10.1126/science.abd4359 . |
| [1] | Min LIANG, Yang GUO, Jinjin WANG, Mengyan ZHU, Jun CHI, Yanjuan CHEN, Chengji WANG, Zhilan YU, Ruling SHEN. Construction of Dmd Gene Mutant Mice and Phenotype Verification in Muscle and Immune Systems [J]. Laboratory Animal and Comparative Medicine, 2024, 44(1): 42-51. |
| [2] | LAI Suomei, DING Yifu, LI Jinsong. A New Strategy for Constructing Mouse Models of Complex Diseases: Semi-cloning Technology Based on Sperm-like Haploid Embryonic Stem Cells [J]. Laboratory Animal and Comparative Medicine, 2021, 41(5): 369-383. |
| [3] | HONG Shenghui, ZHANG Xuliang, WANG Qianqian, LIU Ping, LIU Diwen. Construction of Inhibin Gene Knockout Mice and Preliminary Analysis of the Phenotype [J]. Laboratory Animal and Comparative Medicine, 2020, 40(4): 306-. |
| [4] | GAO Meng-qiao, AI Dong-xu, LI Yu, SUN Fei, WANG Jin, FAN Jun-wen, YUAN Zheng, LIU Yuan, SUN Zhao-zeng. The Construction of Ring Finger Protein 126 Gene Knockout Mouse by Using CRISPR/Cas9 Technique [J]. Laboratory Animal and Comparative Medicine, 2019, 39(1): 21-25. |
| [5] | ZHANG Qi, WANG Jian-fei. Progress and Perspective for Xenotransplantation [J]. Laboratory Animal and Comparative Medicine, 2018, 38(6): 407-411. |
| [6] | AI Dong-xu, ZHONG De-gang, SUN Fei, Li Yu, LI Chong, QIN Tong-tong, GAO Meng-qiao, DONG Shi-shi, SUN Zhao-zeng, LI Lian-rui. Construction of Lyst Gene Defective C57BL/6 Mice by CRISPR/Cas9 Technology [J]. Laboratory Animal and Comparative Medicine, 2018, 38(3): 202-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||