
Laboratory Animal and Comparative Medicine ›› 2025, Vol. 45 ›› Issue (6): 676-687.DOI: 10.12300/j.issn.1674-5817.2025.112
• Invertebrate Laboratory Animal: Fruit fly • Previous Articles Next Articles
Received:2025-07-07
Revised:2025-08-12
Online:2025-12-25
Published:2025-12-19
Contact:
WANG Lu
CLC Number:
WANG Ye,WANG Lu. Drosophila melanogaster Transposons: Characterization, Regulation, and Their Role in Genome Evolution[J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 676-687. DOI: 10.12300/j.issn.1674-5817.2025.112.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.slarc.org.cn/dwyx/EN/10.12300/j.issn.1674-5817.2025.112
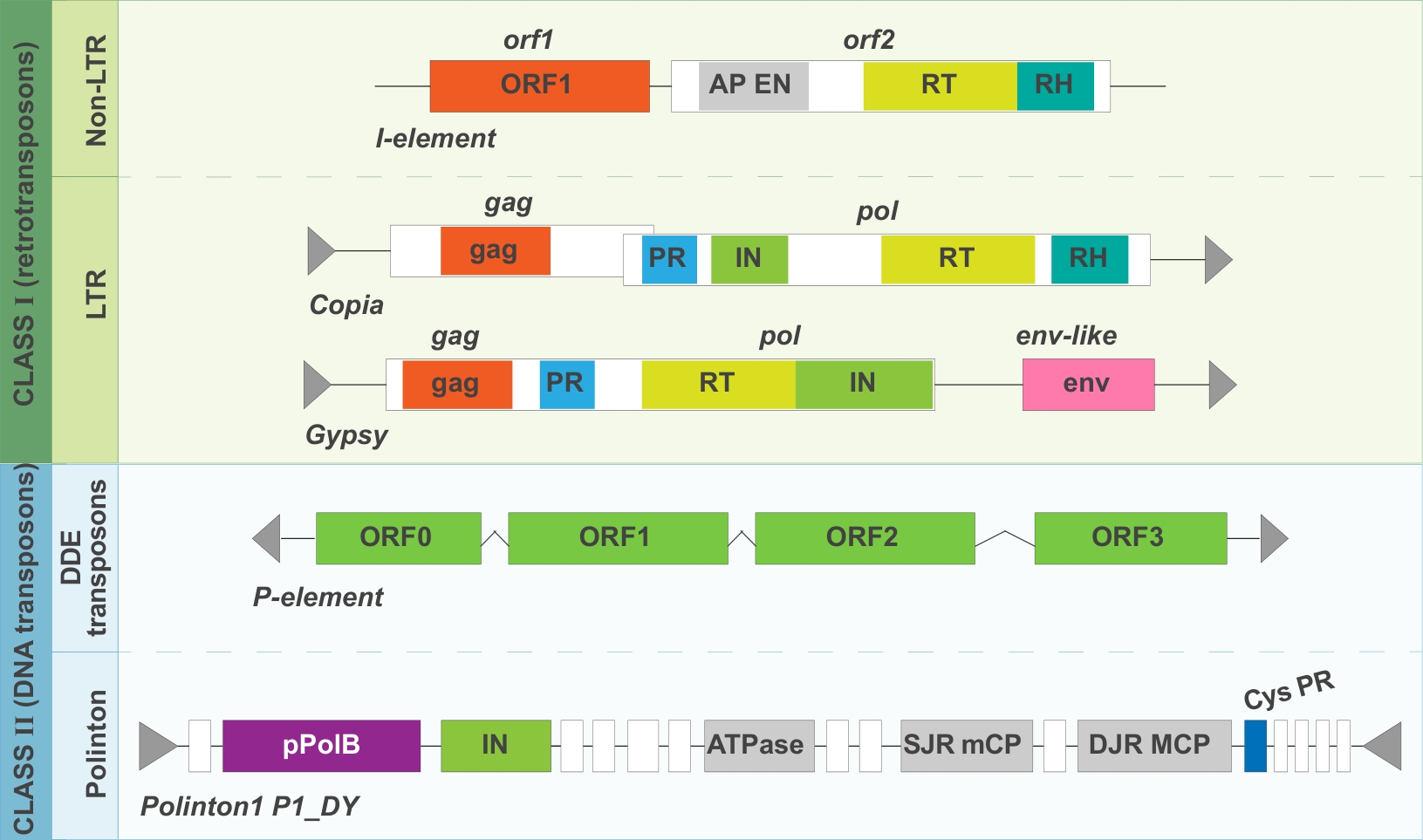
Figure 1 Examples of the structure and classification of Drosophila transposable elementsNote: Black boxes indicate open reading frames; Colored regions indicate defined protein structural domains; Folded segments indicate introns; Triangles indicate repeated sequences; Structural domains of the same color (excluding gray) share a common ancestor; AP, apurinic/apyrimidinic; ATPase, adenosine triphosphatase; Cys, cysteine; DJR MCP, double jelly-roll major capsid protein; EN, endonuclease; IN, integrase; ORF, open reading frame; pPolB, protein-primed type B DNA polymerase; PR, pol-encoded protease; RH, Ribonuclease H domain; RT, reverse transcriptase; SJR mCP, single jelly-roll minor capsid protein.
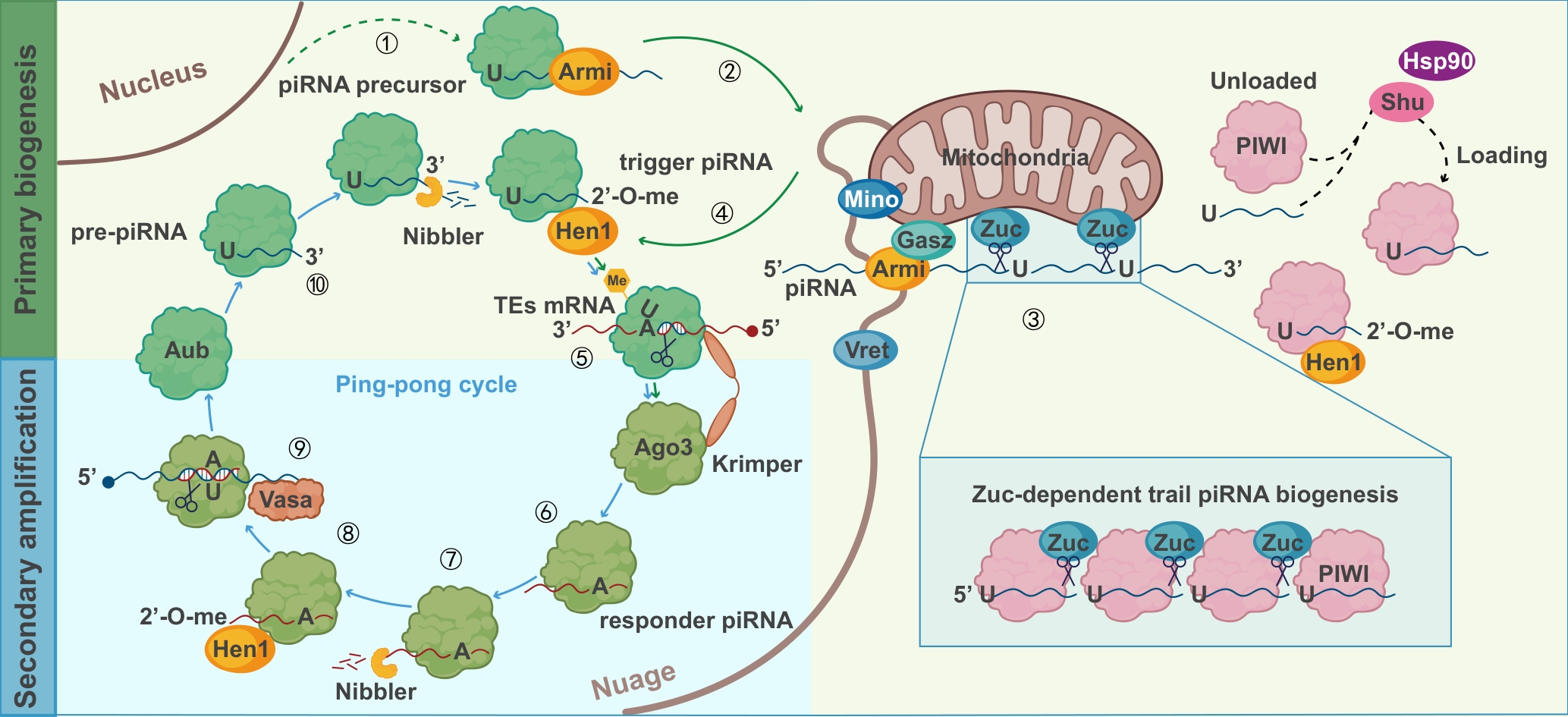
Figure 2 Schematic diagram of piRNA biogenesisNote: 2'-O-me, 2'-O-methylation; Me, methyl group; piRNA, PIWI-interacting RNA; TE, transposable element; Ago3, Argonaute 3; Armi, Armitage; Aub, Aubergine; Hsp90, Heat shock protein 90; Mino, Minotaur; Vret, Vreteno; Zuc, Zucchini; Hen1, Hen1 methyl-transferase; Vasa, Vasa DEAD-box RNA helicase; Gasz, Germline-associated scaffolding zinc finger.
驯化类型 Type of co-option | 驯化机制/来源 Mechanism/Source of co-option | 功能影响 Functional impact | 代表性案例与功能 Representative case & function |
|---|---|---|---|
调控元件驯化 Regulatory element co-option | TEs调控序列(如启动子、增强子、绝缘子) | 改变邻近/宿主基因的表达模式、组织特异性、发育时序或环境响应性 | 果蝇体色演化:TEs(尤其LTR型)插入影响色素相关基因的调控,驱动体色模式快速变化[ |
编码序列驯化(完全蛋白) Protein-coding sequence co-option (full-length) | TEs完整或近乎完整的编码序列 | 产生具有细胞功能的全新结构蛋白或酶 | PIF-like基因:果蝇属中多次独立驯化自PIF转座酶,形成新的功能基因家族[ |
编码序列驯化(功能域/模块) Protein-coding sequence co-option (functional domain/module) | TEs编码蛋白的特定结构域 | 赋予宿主蛋白新功能或参与新细胞过程 | Arc/Arc1蛋白(gag域):来源于逆转录TEs(如Gypsy、Copia)GAG蛋白,形成衣壳样结构,包装自身mRNA,介导神经元-肌肉或神经元间RNA运输,对突触可塑性和记忆维持至关重要[ |
整体元件利用 Holistic element utilization | 特定TEs家族的活性或产物 | 在特定生理或发育过程中被宿主利用 | mdg4(Gypsy家族):在果蝇蛹期变态窗口期激活,通过STING–Relish通路诱导全身性抗病毒免疫状态[ |
Table 1 Examples of major types, mechanisms and functional effects of TEs host co-option in Drosophila
驯化类型 Type of co-option | 驯化机制/来源 Mechanism/Source of co-option | 功能影响 Functional impact | 代表性案例与功能 Representative case & function |
|---|---|---|---|
调控元件驯化 Regulatory element co-option | TEs调控序列(如启动子、增强子、绝缘子) | 改变邻近/宿主基因的表达模式、组织特异性、发育时序或环境响应性 | 果蝇体色演化:TEs(尤其LTR型)插入影响色素相关基因的调控,驱动体色模式快速变化[ |
编码序列驯化(完全蛋白) Protein-coding sequence co-option (full-length) | TEs完整或近乎完整的编码序列 | 产生具有细胞功能的全新结构蛋白或酶 | PIF-like基因:果蝇属中多次独立驯化自PIF转座酶,形成新的功能基因家族[ |
编码序列驯化(功能域/模块) Protein-coding sequence co-option (functional domain/module) | TEs编码蛋白的特定结构域 | 赋予宿主蛋白新功能或参与新细胞过程 | Arc/Arc1蛋白(gag域):来源于逆转录TEs(如Gypsy、Copia)GAG蛋白,形成衣壳样结构,包装自身mRNA,介导神经元-肌肉或神经元间RNA运输,对突触可塑性和记忆维持至关重要[ |
整体元件利用 Holistic element utilization | 特定TEs家族的活性或产物 | 在特定生理或发育过程中被宿主利用 | mdg4(Gypsy家族):在果蝇蛹期变态窗口期激活,通过STING–Relish通路诱导全身性抗病毒免疫状态[ |
| [1] | MCCLINTOCK B. The origin and behavior of mutable loci in maize[J]. Proc Natl Acad Sci USA, 1950, 36(6):344-355. DOI: 10.1073/pnas.36.6.344 . |
| [2] | WELLS J N, FESCHOTTE C. A field guide to eukaryotic transposable elements[J]. Annu Rev Genet, 2020, 54:539-561. DOI: 10.1146/annurev-genet-040620-022145 . |
| [3] | BHAT A, GHATAGE T, BHAN S, et al. Role of transposable elements in genome stability: implications for health and disease[J]. Int J Mol Sci, 2022, 23(14):7802. DOI: 10.3390/ijms23147802 . |
| [4] | LIANG Y H, QU X, SHAH N M, et al. Towards targeting transposable elements for cancer therapy[J]. Nat Rev Cancer, 2024, 24(2):123-140. DOI: 10.1038/s41568-023-00653-8 . |
| [5] | KAZAZIAN H H Jr, MORAN J V. Mobile DNA in health and disease[J]. N Engl J Med, 2017, 377(4):361-370. DOI: 10.1056/NEJMra1510092 . |
| [6] | BIER E. Gene drives gaining speed[J]. Nat Rev Genet, 2022, 23(1):5-22. DOI: 10.1038/s41576-021-00386-0 . |
| [7] | KLEIN S J, O'NEILL R J. Transposable elements: genome innovation, chromosome diversity, and centromere conflict[J]. Chromosome Res, 2018, 26(1-2):5-23. DOI: 10.1007/s10577-017-9569-5 . |
| [8] | DRONGITIS D, ANIELLO F, FUCCI L, et al. Roles of transposable elements in the different layers of gene expression regulation[J]. Int J Mol Sci, 2019, 20(22):5755. DOI: 10.3390/ijms20225755 . |
| [9] | GASPAROTTO E, BURATTIN F V, DI GIOIA V, et al. Transposable elements co-option in genome evolution and gene regulation[J]. Int J Mol Sci, 2023, 24(3):2610. DOI: 10.3390/ijms24032610 . |
| [10] | NISHIHARA H. Transposable elements as genetic accelerators of evolution: contribution to genome size, gene regulatory network rewiring and morphological innovation[J]. Genes Genet Syst, 2020, 94(6):269-281. DOI: 10.1266/ggs.19-00029 . |
| [11] | COSBY R L, CHANG N C, FESCHOTTE C. Host-transposon interactions: conflict, cooperation, and cooption[J]. Genes Dev, 2019, 33(17-18):1098-1116. DOI: 10.1101/gad.327312.119 . |
| [12] | GORBUNOVA V, SELUANOV A, MITA P, et al. The role of retrotransposable elements in ageing and age-associated diseases[J]. Nature, 2021, 596(7870):43-53. DOI: 10.1038/s41586-021-03542-y . |
| [13] | SUN W Y, SAMIMI H, GAMEZ M, et al. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies[J]. Nat Neurosci, 2018, 21(8):1038-1048. DOI: 10.1038/s41593-018-0194-1 . |
| [14] | DELLA VALLE F, REDDY P, AGUIRRE VAZQUEZ A, et al. Reactivation of retrotransposable elements is associated with environmental stress and ageing[J]. Nat Rev Genet, 2025, 26(8):547-558. DOI: 10.1038/s41576-025-00829-y . |
| [15] | LI W H, PRAZAK L, CHATTERJEE N, et al. Activation of transposable elements during aging and neuronal decline in Drosophila [J]. Nat Neurosci, 2013, 16(5):529-531. DOI: 10.1038/nn.3368 . |
| [16] | SCHRADER L, SCHMITZ J. The impact of transposable elements in adaptive evolution[J]. Mol Ecol, 2019, 28(6):1537-1549. DOI: 10.1111/mec.14794 . |
| [17] | HSU P S, YU S H, TSAI Y T, et al. More than causing (epi)genomic instability: emerging physiological implications of transposable element modulation[J]. J Biomed Sci, 2021, 28(1):58. DOI: 10.1186/s12929-021-00754-2 . |
| [18] | GALBRAITH J D, HAYWARD A. The influence of transposable elements on animal colouration[J]. Trends Genet, 2023, 39(8):624-638. DOI: 10.1016/j.tig.2023.04.005 . |
| [19] | DOPKINS N, O'MARA M M, LAWRENCE E, et al. A field guide to endogenous retrovirus regulatory networks[J]. Mol Cell, 2022, 82(20):3763-3768. DOI: 10.1016/j.molcel.2022.09.011 . |
| [20] | CASOLA C, LAWING A M, BETRÁN E, et al. PIF-like transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes[J]. Mol Biol Evol, 2007, 24(8):1872-1888. DOI: 10.1093/molbev/msm116 . |
| [21] | BARRÓN M G, FISTON-LAVIER A S, PETROV D A, et al. Population genomics of transposable elements in Drosophila [J]. Annu Rev Genet, 2014, 48:561-581. DOI: 10.1146/annurev-genet-120213-092359 . |
| [22] | RECH G E, RADÍO S, GUIRAO-RICO S, et al. Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila [J]. Nat Commun, 2022, 13(1):1948. DOI: 10.1038/s41467-022-29518-8 . |
| [23] | SHEN D, XU Y Q, SHI Q, et al. Retrotransposon 3S18 forms self-protective aggregates and prolongs mid-oogenesis[J]. Cell Rep, 2025, 44(7):115914. DOI: 10.1016/j.celrep.2025.115914 . |
| [24] | WANG L, DOU K, MOON S, et al. Hijacking oogenesis enables massive propagation of LINE and retroviral transposons[J]. Cell, 2018, 174(5):1082-1094.e12. DOI: 10.1016/j.cell.2018.06.040 . |
| [25] | MOON S, CASSANI M, LIN Y A, et al. A robust transposon-endogenizing response from germline stem cells[J]. Dev Cell, 2018, 47(5):660-671.e3. DOI: 10.1016/j.devcel.2018.10.011 . |
| [26] | YANG F, SU W J, CHUNG O W, et al. Retrotransposons hijack alt-EJ for DNA replication and eccDNA biogenesis[J]. Nature, 2023, 620(7972):218-225. DOI: 10.1038/s41586-023-06327-7 . |
| [27] | MÉREL V, BOULESTEIX M, FABLET M, et al. Transposable elements in Drosophila [J]. Mob DNA, 2020, 11:23. DOI: 10.1186/s13100-020-00213-z . |
| [28] | SPRADLING A C, RUBIN G M. Transposition of cloned P elements into Drosophila germ line chromosomes[J]. Science, 1982, 218(4570):341-347. DOI: 10.1126/science. 6289435 . |
| [29] | RUBIN G M, SPRADLING A C. Genetic transformation of Drosophila with transposable element vectors[J]. Science, 1982, 218(4570):348-353. DOI: 10.1126/science.6289436 . |
| [30] | HO S, THEURKAUF W, RICE N. piRNA-guided transposon silencing and response to stress in Drosophila germline[J]. Viruses, 2024, 16(5):714. DOI: 10.3390/v16050714 . |
| [31] | CZECH B, MUNAFÒ M, CIABRELLI F, et al. piRNA-guided genome defense: from biogenesis to silencing[J]. Annu Rev Genet, 2018, 52:131-157. DOI: 10.1146/annurev-genet-120417-031441 . |
| [32] | WANG X, RAMAT A, SIMONELIG M, et al. Emerging roles and functional mechanisms of PIWI-interacting RNAs[J]. Nat Rev Mol Cell Biol, 2023, 24(2):123-141. DOI: 10.1038/s41580-022-00528-0 . |
| [33] | LUO Y C, HE P, KANRAR N, et al. Maternally inherited siRNAs initiate piRNA cluster formation[J]. Mol Cell, 2023, 83(21):3835-3851.e7. DOI: 10.1016/j.molcel.2023.09.033 . |
| [34] | LEE Y C G, KARPEN G H. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution[J]. eLife, 2017, 6: e25762. DOI: 10.7554/eLife.25762 . |
| [35] | LEE Y C G. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster [J]. PLoS Genet, 2015, 11(6): e1005269. DOI: 10.1371/journal.pgen.1005269 . |
| [36] | SELVARAJU D, WIERZBICKI F, KOFLER R. Experimentally evolving Drosophila erecta populations may fail to establish an effective piRNA-based host defense against invading P-elements [J]. Genome Res, 2024, 34(3):410-425. DOI: 10.1101/gr.278706.123 . |
| [37] | SATYAKI P V, CUYKENDALL T N, WEI K H, et al. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats[J]. PLoS Genet, 2014, 10(3): e1004240. DOI: 10.1371/journal.pgen.1004240 . |
| [38] | SHIBA T, SAIGO K. Retrovirus-like particles containing RNA homologous to the transposable element Copia in Drosophila melanogaster [J]. Nature, 1983, 302(5904):119-124. DOI: 10.1038/302119a0 . |
| [39] | TOURET F, GUIGUEN F, GREENLAND T, et al. In between: Gypsy in Drosophila melanogaster reveals new insights into endogenous retrovirus evolution[J]. Viruses, 2014, 6(12):4914-4925. DOI: 10.3390/v6124914 . |
| [40] | MOSCHETTI R, DIMITRI P, CAIZZI R, et al. Genomic instability of I elements of Drosophila melanogaster in absence of dysgenic crosses[J]. PLoS One, 2010, 5(10): e13142. DOI: 10.1371/journal.pone.0013142 . |
| [41] | JAILLET J, GENTY M, CAMBEFORT J, et al. Regulation of mariner transposition: the peculiar case of Mos1[J]. PLoS One, 2012, 7(8): e43365. DOI: 10.1371/journal.pone.0043365 . |
| [42] | CAO J C, YU T X, XU B, et al. Epigenetic and chromosomal features drive transposon insertion in Drosophila melanogaster [J]. Nucleic Acids Res, 2023, 51(5):2066-2086. DOI: 10.1093/nar/gkad054 . |
| [43] | LOUBALOVA Z, KONSTANTINIDOU P, HAASE A D. Themes and variations on piRNA-guided transposon control[J]. Mob DNA, 2023, 14(1):10. DOI: 10.1186/s13100-023-00298-2 . |
| [44] | ILIK İ A, YANG X, ZHANG Z Z Z, et al. Transcriptional and post-transcriptional regulation of transposable elements and their roles in development and disease[J]. Nat Rev Mol Cell Biol, 2025, 26(10):759-775. DOI: 10.1038/s41580-025-00867-8 . |
| [45] | JOURAVLEVA K, ZAMORE P D. A guide to the biogenesis and functions of endogenous small non-coding RNAs in animals[J]. Nat Rev Mol Cell Biol, 2025, 26(5):347-370. DOI: 10.1038/s41580-024-00818-9 . |
| [46] | SRIVASTAV S P, FESCHOTTE C, CLARK A G. Rapid evolution of piRNA clusters in the Drosophila melanogaster ovary[J]. Genome Res, 2024, 34(5):711-724. DOI: 10.1101/gr.278062.123 . |
| [47] | PANE A, JIANG P, ZHAO D Y, et al. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline[J]. EMBO J, 2011, 30(22):4601-4615. DOI: 10.1038/emboj.2011.334 . |
| [48] | SUYAMA R, KAI T. piRNA processing within non-membrane structures is governed by constituent proteins and their functional motifs[J]. FEBS J, 2025, 292(11):2715-2736. DOI: 10.1111/febs.17360 . |
| [49] | WU P H, ZAMORE P D. Defining the functions of PIWI-interacting RNAs[J]. Nat Rev Mol Cell Biol, 2021, 22(4):239-240. DOI: 10.1038/s41580-021-00336-y . |
| [50] | CHARY S, HAYASHI R. The absence of core piRNA biogenesis factors does not impact efficient transposon silencing in Drosophila [J]. PLoS Biol, 2023, 21(6): e3002099. DOI: 10.1371/journal.pbio.3002099 . |
| [51] | PODVALNAYA N, BRONKHORST A W, LICHTENBERGER R, et al. piRNA processing by a trimeric Schlafen-domain nuclease[J]. Nature, 2023, 622(7982):402-409. DOI: 10.1038/s41586-023-06588-2 . |
| [52] | DE BRITO T F, ARRUDA CARDOSO M, ATINBAYEVA N, et al. Embryonic piRNAs target horizontally transferred vertebrate transposons in assassin bugs[J]. Front Cell Dev Biol, 2024, 12:1481881. DOI: 10.3389/fcell.2024.1481881 . |
| [53] | VAN LOPIK J, ALIZADA A, TRAPOTSI M A, et al. Unistrand piRNA clusters are an evolutionarily conserved mechanism to suppress endogenous retroviruses across the Drosophila genus[J]. Nat Commun, 2023, 14(1):7337. DOI: 10.1038/s41467-023-42787-1 . |
| [54] | JONES B C, WOOD J G, CHANG C Y, et al. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan[J]. Nat Commun, 2016, 7:13856. DOI: 10.1038/ncomms13856 . |
| [55] | LEWIS S H, QUARLES K A, YANG Y J, et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements[J]. Nat Ecol Evol, 2018, 2(1):174-181. DOI: 10.1038/s41559-017-0403-4 . |
| [56] | YU T X, BLYTON M B J, ABAJORGA M, et al. Evolution of KoRV-a transcriptional silencing in wild koalas[J]. Cell, 2025, 188(8):2081-2093.e16. DOI: 10.1016/j.cell.2025.02.006 . |
| [57] | YU T X, KOPPETSCH B S, PAGLIARANI S, et al. The piRNA response to retroviral invasion of the koala genome[J]. Cell, 2019, 179(3):632-643.e12. DOI: 10.1016/j.cell.2019.09.002 . |
| [58] | GEBERT D, NEUBERT L K, LLOYD C, et al. Large Drosophila germline piRNA clusters are evolutionarily labile and dispensable for transposon regulation[J]. Mol Cell, 2021, 81(19):3965-3978.e5. DOI: 10.1016/j.molcel.2021.07.011 . |
| [59] | WOOD J G, JONES B C, JIANG N, et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila [J]. Proc Natl Acad Sci USA, 2016, 113(40):11277-11282. DOI: 10.1073/pnas.1604621113 . |
| [60] | ZHANG G, YU T X, PARHAD S S, et al. piRNA-independent transposon silencing by the Drosophila THO complex[J]. Dev Cell, 2021, 56(18):2623-2635.e5. DOI: 10.1016/j.devcel.2021. 08.021 . |
| [61] | ZATTERA M L, BRUSCHI D P. Transposable elements as a source of novel repetitive DNA in the eukaryote genome[J]. Cells, 2022, 11(21):3373. DOI: 10.3390/cells11213373 . |
| [62] | CASACUBERTA E. Drosophila: retrotransposons making up telomeres[J]. Viruses, 2017, 9(7):192. DOI: 10.3390/v9070192 . |
| [63] | LEVIS R W, GANESAN R, HOUTCHENS K, et al. Transposons in place of telomeric repeats at a Drosophila telomere[J]. Cell, 1993, 75(6):1083-1093. DOI: 10.1016/0092-8674(93)90318-k . |
| [64] | SONG J M, LIU J X, SCHNAKENBERG S L, et al. Variation in piRNA and transposable element content in strains of Drosophila melanogaster [J]. Genome Biol Evol, 2014, 6(10):2786-2798. DOI: 10.1093/gbe/evu217 . |
| [65] | WIDEN S A, BES I C, KORESHOVA A, et al. Virus-like transposons cross the species barrier and drive the evolution of genetic incompatibilities[J]. Science, 2023, 380(6652): eade0705. DOI: 10.1126/science.ade0705 . |
| [66] | SCHWARZ F, WIERZBICKI F, SENTI K A, et al. Tirant stealthily invaded natural Drosophila melanogaster populations during the last century[J]. Mol Biol Evol, 2021, 38(4):1482-1497. DOI: 10.1093/molbev/msaa308 . |
| [67] | PASTUZYN E D, DAY C E, KEARNS R B, et al. The neuronal gene Arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer[J]. Cell, 2018, 172(1-2):275-288.e18. DOI: 10.1016/j.cell.2017.12.024 . |
| [68] | ASHLEY J, CORDY B, LUCIA D, et al. Retrovirus-like gag protein Arc1 binds RNA and traffics across synaptic boutons[J]. Cell, 2018, 172(1-2):262-274.e11. DOI: 10.1016/j.cell.2017. 12.022 . |
| [69] | WANG L, TRACY L, SU W J, et al. Retrotransposon activation during Drosophila metamorphosis conditions adult antiviral responses[J]. Nat Genet, 2022, 54(12):1933-1945. DOI: 10.1038/s41588-022-01214-9 . |
| [70] | CHANG Y H, KEEGAN R M, PRAZAK L, et al. Cellular labeling of endogenous retrovirus replication (CLEVR) reveals de novo insertions of the gypsy retrotransposable element in cell culture and in both neurons and glial cells of aging fruit flies[J]. PLoS Biol, 2019, 17(5): e3000278. DOI: 10.1371/journal.pbio.3000278 . |
| [71] | HOU Y Z, LI Y P, XIANG J F, et al. TDP-43 chronic deficiency leads to dysregulation of transposable elements and gene expression by affecting R-loop and 5hmC crosstalk[J]. Cell Rep, 2024, 43(1):113662. DOI: 10.1016/j.celrep.2023.113662 . |
| [72] | KRUG L, CHATTERJEE N, BORGES-MONROY R, et al. Retrotransposon activation contributes to neurode-generation in a Drosophila TDP-43 model of ALS[J]. PLoS Genet, 2017, 13(3): e1006635. DOI: 10.1371/journal.pgen. 1006635 . |
| [73] | SONG S U, GERASIMOVA T, KURKULOS M, et al. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus[J]. Genes Dev, 1994, 8(17):2046-2057. DOI: 10.1101/gad.8.17.2046 . |
| [74] | YOTH M, MAUPETIT-MÉHOUAS S, AKKOUCHE A, et al. Reactivation of a somatic errantivirus and germline invasion in Drosophila ovaries[J]. Nat Commun, 2023, 14(1):6096. DOI: 10.1038/s41467-023-41733-5 . |
| [75] | KLUMPE S, SENTI K A, BECK F, et al. In-cell structure and snapshots of Copia retrotransposons in intact tissue by cryo-ET[J]. Cell, 2025, 188(8):2094-2110.e18. DOI: 10.1016/j.cell.2025.02.003 . |
| [76] | PIETZENUK B, MARKUS C, GAUBERT H, et al. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements[J]. Genome Biol, 2016, 17(1):209. DOI: 10.1186/s13059-016-1072-3 . |
| [1] | WANG Hanyue, CHEN Jiawei, GAO Xiangbin, LUO Wei, LIU Suning. Research Overview on Corpora Cardiaca Function of Drosophila melanogaster [J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 705-718. |
| [2] | WANG Mingzhu, GAO Yinghao, TAN Shuangshuang, WU Wei. Construction and Characterization of UAS-Irk3-EGFP Transgenic Drosophila Lines [J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 656-662. |
| [3] | CHEN Haotian, LIU Jingnan. Applications and Advances of Drosophila in Research of Obesity and Its Related Metabolic Diseases [J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 688-704. |
| [4] | DENG Xianming, WANG Fei. Research Progress on Drosophila Electron Microscopy Connectome Database and Functional Analysis of Related Neural Circuits [J]. Laboratory Animal and Comparative Medicine, 2025, 45(6): 663-675. |
| [5] | Longmei XU, Ruling SHEN, Chun FAN, Wei WU. Generation of 12 Drosophila Transgenic Negative Control Lines Based on Site-specific ΦC31 Integrase and pUASTattB Vector [J]. Laboratory Animal and Comparative Medicine, 2023, 43(5): 541-547. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

